diamond大基因序列快速比对工具使用详解-包含超算集群多节点计算使用方法
Diamond是一款快速的序列比对工具,其使用方法如下:
1. 安装Diamond:
可从官方网站(https://github.com/bbuchfink/diamond/releases)下载安装包,并安装到本地电脑中。当然还有docker,conda以及编译安装方式,一般用不上,但注意新版对gcc的要求高,出现gcc错误时可选择下载低版本的diamond或者升级gcc到指定版本以上。
#下载diamond程序文件
wget http://github.com/bbuchfink/diamond/releases/download/v2.1.8/diamond-linux64.tar.gz
###其他版本直接访问http://github.com/bbuchfink/diamond/releases/download/查看#解压会出来一个diamond的文件
tar -xzvf diamond-linux64.tar.gz
#移到系统环境目录、或将当前目录加入系统环境目录,或者直接使用路径加diamond命令运行
diamond blastx./diamond blastx/opt/diamond blastx2. 准备数据集:
首先需要准备用于比对的序列数据集,比如fasta格式的序列文件。
#下载nr数据库,或这自己需要的数据库
wget ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz
gunzip nr.gz
#使用diamond命令创建dimond格式数据库
diamond makedb --in nr --db nr3. 运行Diamond:
常规使用
在终端中输入以下命令,即可启动Diamond程序并运行比对任务:
diamond blastx -d [参考序列文件] -q [待比对序列文件] -o [输出文件名]
#下载nr数据库,或这自己需要的数据库
wget ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz
gunzip nr.gz
#使用diamond命令创建dimond格式数据库
diamond makedb --in nr --db nr#命令使用
diamond blastx --db nr -q reads.fna -o dna_matches_fmt6.txt
diamond blastp --db nr -q reads.faa -o protein_matches_fmt6.txt
其中,blastx表示使用蛋白质序列比对算法,-d和-q分别指定参考序列文件和待比对序列文件,-o指定输出文件名。
超算集群多计算节点并行计算(私房菜)Distributed computing
diamond尽管速度快,但对于大文件进行比对时,大于1G以上的文件对于40核的单个节点可能仍然需要几天的时间,如果有较多的节点时,可以使用多节点的并行计算,这一点太给力了。
准备工作(重要,否则不成功):
1、将diamond程序目录在各节点间共享
2、样品序列目录在各节点间共享
3、所有节点使用相同的临时目录在各节点间共享。
# Diamond distributed-memory parallel processing
#Diamond supports the parallel processing of large alignments on HPC clusters and #supercomputers, spanning numerous compute nodes. Work distribution is orchestrated by #Diamond via a shared file system typically available on such clusters, using lightweight #file-based stacks and POSIX functionality.#Usage
#To run Diamond in parallel, two steps need to be performed. First, during a short #initialization run using a single process, the query and database are scanned and chunks #of work are written to the file-based stacks on the parallel file system. Second, the #actual parallel run is performed, where multiple DIAMOND processes on different compute #nodes work on chunks of the query and the reference database to perform alignments and #joins.#Initialization 先进行任务初始化,这个只需要在第一个节点上初始化就行了。其他节点直接启动后面一步的并行计算命令就行
#The initialization of a parallel run should be done (e.g. interactively on a login node) #using the parameters --multiprocessing --mp-init as follows:diamond blastp --db DATABASE.dmnd --query QUERY.fasta --multiprocessing --mp-init --tmpdir $TMPDIR --parallel-tmpdir $PTMPDIR#Here $TMPDIR refers to a local temporary directory, whereas $PTMPDIR refers to a #directory in the parallel file system where the file-based stacks containing the work #packages will be created. Note that the size of the chunking and thereby the number of #work packages is controlled via the --block-size parameter.#Parallel run 开始真实的并行计算,可以在所有计算节点启动
#The actual parallel run should be done using the parameter --multiprocessing as follows:diamond blastp --db DATABASE.dmnd --query QUERY.fasta -o OUTPUT_FILE --multiprocessing --tmpdir $TMPDIR --parallel-tmpdir $PTMPDIR#这里特意说明文件夹与任务初始化文件夹的一致性,主要是临时计算目录tmpdir
#Note that $PTMPDIR must refer to the same location as used during the initialization, #and it must be accessible from any of the compute nodes involved. To launch the parallel #processes on many nodes, a batch system such as SLURM is typically used. For the output #not a single stream is used but rather multiple files are created, one for each query #chunk.#SLURM batch file example slurm超算集群脚本,这个不多说了吧,使用这个更方便一点,没有也不用担心,使用前面那两个命令即可。
#The following script shows an example of how a massively parallel can be performed using #the SLURM batch system on a supercomputer.#!/bin/bash -l
#SBATCH -D ./
#SBATCH -J DIAMOND
#SBATCH --mem=185000
#SBATCH --nodes=520
#SBATCH --ntasks-per-node=1
#SBATCH --ntasks-per-core=2
#SBATCH --cpus-per-task=80
#SBATCH --mail-type=none
#SBATCH --time=24:00:00module purge
module load gcc impi
export SLURM_HINT=multithread###以下是超算的相关说明,重点关注前面配置即可。
srun diamond FLAGS
FLAGS refers to the aforementioned parallel flags for Diamond. Note that the actual configuration of the nodes varies between machines, and therefore, the parameters shown here are not of general applicability. It is recommended to start with few nodes on small problems, first.Abort and resume
Parallel runs can be aborted and later resumed, and unfinished work packages from a previous run can be recovered and resubmitted in a subsequent run.Using the option --multiprocessing --mp-recover for the same value of --parallel-tmpdir will scan the working directory and configure a new parallel run including only the work packages that have not been completed in the previous run.Placing a file stop in the working directory causes DIAMOND processes to shut down in a controlled way after finishing the current work package. After removing the stop file, the multiprocessing run can be continued.Parameter optimization
The granularity of the size of the work packages can be adjusted via the --block-size which at the same time affects the memory requirements at runtime. Parallel runs on more than 512 nodes of a supercomputer have been performed successfully.4. 结果解读:
比对结束后,可以查看输出文件中的比对结果。Diamond的输出文件包含每个待比对序列的匹配结果,包括匹配的参考序列名、匹配位置、匹配得分等信息。
结果字段表示与原生blast结果表示相同:
见: 生物信息学分析-blast序列比对及结果详细说明-CSDN博客
5.帮助说明
以上就是Diamond的基本使用方法,更详细的说明可以参考官方文档:https://github.com/bbuchfink/diamond/wiki。
# downloading the tool,下载工具
wget http://github.com/bbuchfink/diamond/releases/download/v2.1.8/diamond-linux64.tar.gz
tar xzf diamond-linux64.tar.gz
# creating a diamond-formatted database file 创建diamond数据库
./diamond makedb --in reference.fasta -d reference
# running a search in blastp mode 使用blastp模式比对序列
./diamond blastp -d reference -q queries.fasta -o matches.tsv
# running a search in blastx mode 使用blastx 模式比对序列
./diamond blastx -d reference -q reads.fasta -o matches.tsv
# downloading and using a BLAST database
update_blastdb.pl --decompress --blastdb_version 5 swissprot
./diamond prepdb -d swissprot
./diamond blastp -d swissprot -q queries.fasta -o matches.tsvSome important points to consider:Repeat masking is applied to the query and reference sequences by default. To disable it, use --masking 0. 默认情况下是允许重复结果,如果只输出最优结果就加上 --masking 0
DIAMOND is optimized for large input files of >1 million proteins. Naturally the tool can be used for smaller files as well, but the algorithm will not reach its full efficiency.
The program may use quite a lot of memory and also temporary disk space. Should the program fail due to running out of either one, you need to set a lower value for the block size parameter -b. DIAMOND是大文件效率更好,对于小文件建议添加 -b 的参数
The sensitivity can be adjusted using the options --fast, --mid-sensitive, --sensitive, --more-sensitive, --very-sensitive and --ultra-sensitive. 比对敏感性,越往后其结果越接近原生blast结果,但速度也越慢,一般使用--more-sensitive比较适中,计算资源不够的就使用fast。全参数帮助文件
下面是diamond的较为详细的帮助说明:自己慢慢看吧,不过一般不用特意设置了。
diamond --help
diamond v2.0.11.149 (C) Max Planck Society for the Advancement of Science
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)Syntax: diamond COMMAND [OPTIONS]Commands:
makedb Build DIAMOND database from a FASTA file #以fasta文件创建diamond格式数据库
blastp Align amino acid query sequences against a protein reference database #功能与原生blastp功能一致
blastx Align DNA query sequences against a protein reference database #功能与原生blastx一致
view View DIAMOND alignment archive (DAA) formatted file
help Produce help message
version Display version information
getseq Retrieve sequences from a DIAMOND database file
dbinfo Print information about a DIAMOND database file
test Run regression tests
makeidx Make database indexGeneral options:
--threads (-p) number of CPU threads #指定需要运行的线程数,可尽量大
--db (-d) database file #diamond makedb产生的diamond可使用格式的数据库
--out (-o) output file #比对结果输出命名
--outfmt (-f) output format #outfmt,一般选6表格格式,与原生blast一致0 = BLAST pairwise5 = BLAST XML6 = BLAST tabular100 = DIAMOND alignment archive (DAA)101 = SAMValue 6 may be followed by a space-separated list of these keywords:qseqid means Query Seq - idqlen means Query sequence lengthsseqid means Subject Seq - idsallseqid means All subject Seq - id(s), separated by a ';'slen means Subject sequence lengthqstart means Start of alignment in queryqend means End of alignment in querysstart means Start of alignment in subjectsend means End of alignment in subjectqseq means Aligned part of query sequenceqseq_translated means Aligned part of query sequence (translated)full_qseq means Query sequencefull_qseq_mate means Query sequence of the matesseq means Aligned part of subject sequencefull_sseq means Subject sequenceevalue means Expect valuebitscore means Bit scorescore means Raw scorelength means Alignment lengthpident means Percentage of identical matchesnident means Number of identical matchesmismatch means Number of mismatchespositive means Number of positive - scoring matchesgapopen means Number of gap openingsgaps means Total number of gapsppos means Percentage of positive - scoring matchesqframe means Query framebtop means Blast traceback operations(BTOP)cigar means CIGAR stringstaxids means unique Subject Taxonomy ID(s), separated by a ';' (in numerical order)sscinames means unique Subject Scientific Name(s), separated by a ';'sskingdoms means unique Subject Super Kingdom(s), separated by a ';'skingdoms means unique Subject Kingdom(s), separated by a ';'sphylums means unique Subject Phylum(s), separated by a ';'stitle means Subject Titlesalltitles means All Subject Title(s), separated by a '<>'qcovhsp means Query Coverage Per HSPscovhsp means Subject Coverage Per HSPqtitle means Query titleqqual means Query quality values for the aligned part of the queryfull_qqual means Query quality valuesqstrand means Query strandDefault: qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue bitscore
--verbose (-v) verbose console output
--log enable debug log
--quiet disable console output
--header Write header lines to blast tabular format.Makedb options:
--in input reference file in FASTA format
--taxonmap protein accession to taxid mapping file
--taxonnodes taxonomy nodes.dmp from NCBI
--taxonnames taxonomy names.dmp from NCBIAligner options:
--query (-q) input query file
--strand query strands to search (both/minus/plus)
--un file for unaligned queries
--al file or aligned queries
--unfmt format of unaligned query file (fasta/fastq)
--alfmt format of aligned query file (fasta/fastq)
--unal report unaligned queries (0=no, 1=yes)
--max-target-seqs (-k) maximum number of target sequences to report alignments for (default=25)
--top report alignments within this percentage range of top alignment score (overrides --max-target-seqs)
--max-hsps maximum number of HSPs per target sequence to report for each query (default=1)
--range-culling restrict hit culling to overlapping query ranges
--compress compression for output files (0=none, 1=gzip, zstd)
--evalue (-e) maximum e-value to report alignments (default=0.001)
--min-score minimum bit score to report alignments (overrides e-value setting)
--id minimum identity% to report an alignment
--query-cover minimum query cover% to report an alignment
--subject-cover minimum subject cover% to report an alignment
--fast enable fast mode
--mid-sensitive enable mid-sensitive mode
--sensitive enable sensitive mode)
--more-sensitive enable more sensitive mode
--very-sensitive enable very sensitive mode
--ultra-sensitive enable ultra sensitive mode
--iterate iterated search with increasing sensitivity
--global-ranking (-g) number of targets for global ranking
--block-size (-b) sequence block size in billions of letters (default=2.0)
--index-chunks (-c) number of chunks for index processing (default=4)
--tmpdir (-t) directory for temporary files
--parallel-tmpdir directory for temporary files used by multiprocessing
--gapopen gap open penalty
--gapextend gap extension penalty
--frameshift (-F) frame shift penalty (default=disabled)
--long-reads short for --range-culling --top 10 -F 15
--matrix score matrix for protein alignment (default=BLOSUM62)
--custom-matrix file containing custom scoring matrix
--comp-based-stats composition based statistics mode (0-4)
--masking enable tantan masking of repeat regions (0/1=default)
--query-gencode genetic code to use to translate query (see user manual)
--salltitles include full subject titles in DAA file
--sallseqid include all subject ids in DAA file
--no-self-hits suppress reporting of identical self hits
--taxonlist restrict search to list of taxon ids (comma-separated)
--taxon-exclude exclude list of taxon ids (comma-separated)
--seqidlist filter the database by list of accessions
--skip-missing-seqids ignore accessions missing in the databaseAdvanced options:
--algo Seed search algorithm (0=double-indexed/1=query-indexed/ctg=contiguous-seed)
--bin number of query bins for seed search
--min-orf (-l) ignore translated sequences without an open reading frame of at least this length
--freq-sd number of standard deviations for ignoring frequent seeds
--id2 minimum number of identities for stage 1 hit
--xdrop (-x) xdrop for ungapped alignment
--gapped-filter-evalue E-value threshold for gapped filter (auto)
--band band for dynamic programming computation
--shapes (-s) number of seed shapes (default=all available)
--shape-mask seed shapes
--multiprocessing enable distributed-memory parallel processing
--mp-init initialize multiprocessing run
--mp-recover enable continuation of interrupted multiprocessing run
--mp-query-chunk process only a single query chunk as specified
--ext-chunk-size chunk size for adaptive ranking (default=auto)
--no-ranking disable ranking heuristic
--ext Extension mode (banded-fast/banded-slow/full)
--culling-overlap minimum range overlap with higher scoring hit to delete a hit (default=50%)
--taxon-k maximum number of targets to report per species
--range-cover percentage of query range to be covered for range culling (default=50%)
--dbsize effective database size (in letters)
--no-auto-append disable auto appending of DAA and DMND file extensions
--xml-blord-format Use gnl|BL_ORD_ID| style format in XML output
--stop-match-score Set the match score of stop codons against each other.
--tantan-minMaskProb minimum repeat probability for masking (default=0.9)
--file-buffer-size file buffer size in bytes (default=67108864)
--memory-limit (-M) Memory limit for extension stage in GB
--no-unlink Do not unlink temporary files.
--target-indexed Enable target-indexed mode
--ignore-warnings Ignore warningsView options:
--daa (-a) DIAMOND alignment archive (DAA) file
--forwardonly only show alignments of forward strandGetseq options:
--seq Sequence numbers to display.Online documentation at http://www.diamondsearch.org
新版本帮助文件:
新版本帮助更简洁,不在一个层次的命令不显示出来,以免混淆。
diamond --help
diamond v2.1.8.162 (C) Max Planck Society for the Advancement of Science, Benjamin Buchfink, University of Tuebingen
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)Syntax: diamond COMMAND [OPTIONS]Commands:
makedb Build DIAMOND database from a FASTA file
prepdb Prepare BLAST database for use with Diamond
blastp Align amino acid query sequences against a protein reference database
blastx Align DNA query sequences against a protein reference database
cluster Cluster protein sequences
linclust Cluster protein sequences in linear time
realign Realign clustered sequences against their centroids
recluster Recompute clustering to fix errors
reassign Reassign clustered sequences to the closest centroid
view View DIAMOND alignment archive (DAA) formatted file
merge-daa Merge DAA files
help Produce help message
version Display version information
getseq Retrieve sequences from a DIAMOND database file
dbinfo Print information about a DIAMOND database file
test Run regression tests
makeidx Make database index
greedy-vertex-cover Compute greedy vertex coverPossible [OPTIONS] for COMMAND can be seen with syntax: diamond COMMANDOnline documentation at http://www.diamondsearch.org
要显示更具体的命令下的参数,直接增加功能命令回车即可显示,具体使用大家可在自己系统里面查看即可:
diamond makedb
diamond v2.1.8.162 (C) Max Planck Society for the Advancement of Science, Benjamin Buchfink, University of Tuebingen
Documentation, support and updates available at http://www.diamondsearch.org
Please cite: http://dx.doi.org/10.1038/s41592-021-01101-x Nature Methods (2021)Options:
--threads number of CPU threads
--verbose verbose console output
--log enable debug log
--quiet disable console output
--tmpdir directory for temporary files
--db database file
--in input reference file in FASTA format/input DAA files for merge-daa
--taxonmap protein accession to taxid mapping file
--taxonnodes taxonomy nodes.dmp from NCBI
--taxonnames taxonomy names.dmp from NCBI
--file-buffer-size file buffer size in bytes (default=67108864)
--no-unlink Do not unlink temporary files.
--ignore-warnings Ignore warnings
--no-parse-seqids Print raw seqids without parsingError: Missing parameter: database file (--db/-d)
6. 参考文献
Benjamin Buchfink, Chao Xie, and Daniel H. Huson. Fast and sensitive protein alignment using diamond. Nature methods, 12(1):59–60, Jan 2015.
相关文章:

diamond大基因序列快速比对工具使用详解-包含超算集群多节点计算使用方法
Diamond是一款快速的序列比对工具,其使用方法如下: 1. 安装Diamond: 可从官方网站(https://github.com/bbuchfink/diamond/releases)下载安装包,并安装到本地电脑中。当然还有docker,conda以及…...
最新ai系统ChatGPT商业运营版网站源码+支持GPT4.0/支持AI绘画+已支持OpenAI GPT全模型+国内AI全模型+绘画池系统
一、AI创作系统 SparkAi创作系统是基于OpenAI很火的ChatGPT进行开发的Ai智能问答系统和Midjourney绘画系统,支持OpenAI-GPT全模型国内AI全模型。本期针对源码系统整体测试下来非常完美,可以说SparkAi是目前国内一款的ChatGPT对接OpenAI软件系统。那么如…...

ffmpeg x264 x265 fdk-aac 编译记录
ffmpeg 裁剪定制编译过多次了 这里记录下 主题部分 关于ffmpeg自定义编解码器 FFmpeg codec HOWTO - MultimediaWiki 一 环境 ubuntu 18.04 ffmpeg: v4.2.2 ndk :android-ndk-r20b-linux-x86_64 fdk-aac 0.1.5 x264: 0.164.x 1.1 下载编译fdk-aac wget http://ja…...

K8s集群
统一时间:ntpdate(都做) ntpdate -b ntp1.aliyun.com */1 * * * * /usr/sbin/ntpdate -b ntp1.aliyun.com systemctl status docker vi /etc/docker/daemon.json systemctl restart docker m: vim kubernetes.sh cat >> /etc/yum.repos.d/kubernetes.repo…...

生产级 React 框架介绍
文章目录 生产级 React 框架生产级 React 框架Next.jsRemixGatsbyExpo 如何选择生产级 React 框架 生产级 React 框架 React 是一个流行的 JavaScript 框架,用于构建用户界面。React 框架可以帮助你快速构建高质量的 React 应用,但并不是所有的 React 框…...

如何理解 Spring Boot 中的 Starter?
Starter 是 Spring Boot 的四大核心功能特性之一,除此之外,Spring Boot 还有自动装配、Actuator 监控等特性。Spring Boot 里面的这些特性,都是为了让开发者在开发基于 Spring 生态下的企业级应用时,只需要关心业务逻辑,减少对配置…...

vue-query的使用
vue-query,类似于vuex/pinia,以缓存为目的,但侧重的是对网络请求的缓存。 这是我预想的使用场景:假设在各个页面都需要发起相同的请求,去获取数据,而这种数据在一定时间内不会发生变化,那么这种…...
git本地搭建服务器[Vmware虚拟机访问window的git服务器]
先按照https://zhuanlan.zhihu.com/p/494988089说明下载好Gitblit然后复制到tomcat的webapps目录下,如下: 双击"startup.bat"启动tomcat: 然后访问"http://127.0.0.1:8080/gitblit/"即可看到git的界面: 说明git服务器已经能够成功运行了! Vmware虚拟机…...

【Python】基础练习题
1)从random库中选取相应的函数,用蒙特卡罗方法(统计实验方法)求解pi。 import randomdef estimate_pi(num_experiments):num_points_in_circle 0num_total_points 0for _ in range(num_experiments):x random.uniform(-1, 1)y…...

语雀故障与反思,顺便再领半年会员!
23 日语雀的故障相信大部分人都已经知道了,官方发布的公告是这样的: 10 月 23 日语雀出现重大服务故障,且持续 7 个多小时才完全恢复,给用户使用造成极大不便,对此我们深感抱歉。经过复盘,我们在这里向大家…...

PYTHON利用SQLAlchemy库高效操作关联的数据表
SQLAlchemy是一个Python库,用于处理关系型数据库的ORM(对象关系映射)和SQL表达式的生成。它提供了许多功能,包括: ORM(对象关系映射):允许将数据库表映射到Python对象,使…...

TypeScript中的类型工具
类型工具 是ts提供的一些内置的工具,用来更方便地处理各种类型,以及生成新的类型,可以直接使用。也就是对类型的操作。 1. 字符串类型工具 ts 内置了四个字符串类型工具,专门用来操作字符串类型。这四个工具类型都定义在 ts 自…...

File --JAVA
File --JAVA 构造方法 方法说明public File (String pathname)根据文件路径创建对象public File (String parent, String child)根据父路径名字字符串和子路径名字符串创建文件对象public File (String parent, String child)根据父路径对应文件对象和子路径名字符串创建文件…...
比较Excel中的两列目录编号是否一致
使用java代码比较excel中两列是否有包含关系,若有包含关系,核对编号是否一致。 excel数据样例如下: package com.itownet.hg;import org.apache.poi.xssf.usermodel.XSSFSheet; import org.apache.poi.xssf.usermodel.XSSFWorkbook;import j…...

pgsql 创建自增ID , 指定自增ID起始值
1. 创建序列: CREATE SEQUENCE table_name_id_seq; 2. 将序列与表的列关联: ALTER TABLE table_name ALTER COLUMN id SET DEFAULT nextval(table_name_id_seq);3. 设置序列的起始值、递增步长和最大值 // 将序列的起始值设置为 1 ALTER SEQUENCE ta…...

数据安全的重要性:如何解密[thekeyishere@cock.li].Elbie勒索病毒
尊敬的读者: 随着数字时代的来临,网络威胁也不断进化,而[datastorecyberfear.com].Elbie勒索病毒是其中的一个引人注目的例子。这个恶意软件采用高度精密的方法,将用户的数据文件锁定,并要求支付赎金以获取解锁密钥。…...

图像识别在自动驾驶汽车中的决策规划与控制策略研究。
图像识别在自动驾驶汽车中的决策规划与控制策略研究 随着自动驾驶技术的不断发展,图像识别已经成为实现自动驾驶的关键技术之一。在自动驾驶汽车中,图像识别技术主要用于环境感知、决策规划和控制系统。本文将重点探讨图像识别在自动驾驶汽车中的决策规…...

Spring MVC 的责任链模式
Spring MVC 框架使用了责任链模式来处理HTTP请求的流程。这个责任链模式主要包括多个拦截器(Interceptor)以及处理器(Handler),它们协同工作以完成请求的处理和响应。以下是Spring MVC的责任链模式的工作原理和流程图&…...
提升用户体验的关键步骤
快速搭建功能齐全的户外帐篷用具小程序,是现今越来越流行的一种商业模式。通过将线下的户外用品店转移到线上,不仅可以减少人力成本和租金等固定支出,还可以为用户提供更便捷的购物体验。因此,学习如何快速搭建一个功能齐全的户外…...
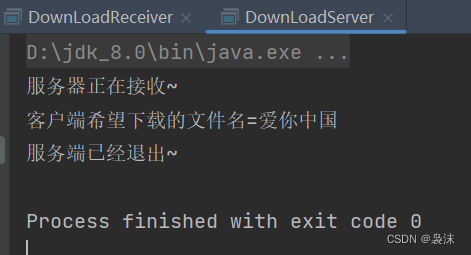
本地模拟,服务器下载文件
题目要求: 编写客户端程序和服务器端程序客户端可以输入一个音乐 文件名,比如 美丽中国,服务端 收到音乐后,可以给客户端返回这个音乐文件,如果服务器没有这个文件,返回一个默认的音乐即可客户端收到文件后…...

k8s从入门到放弃之Ingress七层负载
k8s从入门到放弃之Ingress七层负载 在Kubernetes(简称K8s)中,Ingress是一个API对象,它允许你定义如何从集群外部访问集群内部的服务。Ingress可以提供负载均衡、SSL终结和基于名称的虚拟主机等功能。通过Ingress,你可…...

从零实现富文本编辑器#5-编辑器选区模型的状态结构表达
先前我们总结了浏览器选区模型的交互策略,并且实现了基本的选区操作,还调研了自绘选区的实现。那么相对的,我们还需要设计编辑器的选区表达,也可以称为模型选区。编辑器中应用变更时的操作范围,就是以模型选区为基准来…...

.Net框架,除了EF还有很多很多......
文章目录 1. 引言2. Dapper2.1 概述与设计原理2.2 核心功能与代码示例基本查询多映射查询存储过程调用 2.3 性能优化原理2.4 适用场景 3. NHibernate3.1 概述与架构设计3.2 映射配置示例Fluent映射XML映射 3.3 查询示例HQL查询Criteria APILINQ提供程序 3.4 高级特性3.5 适用场…...
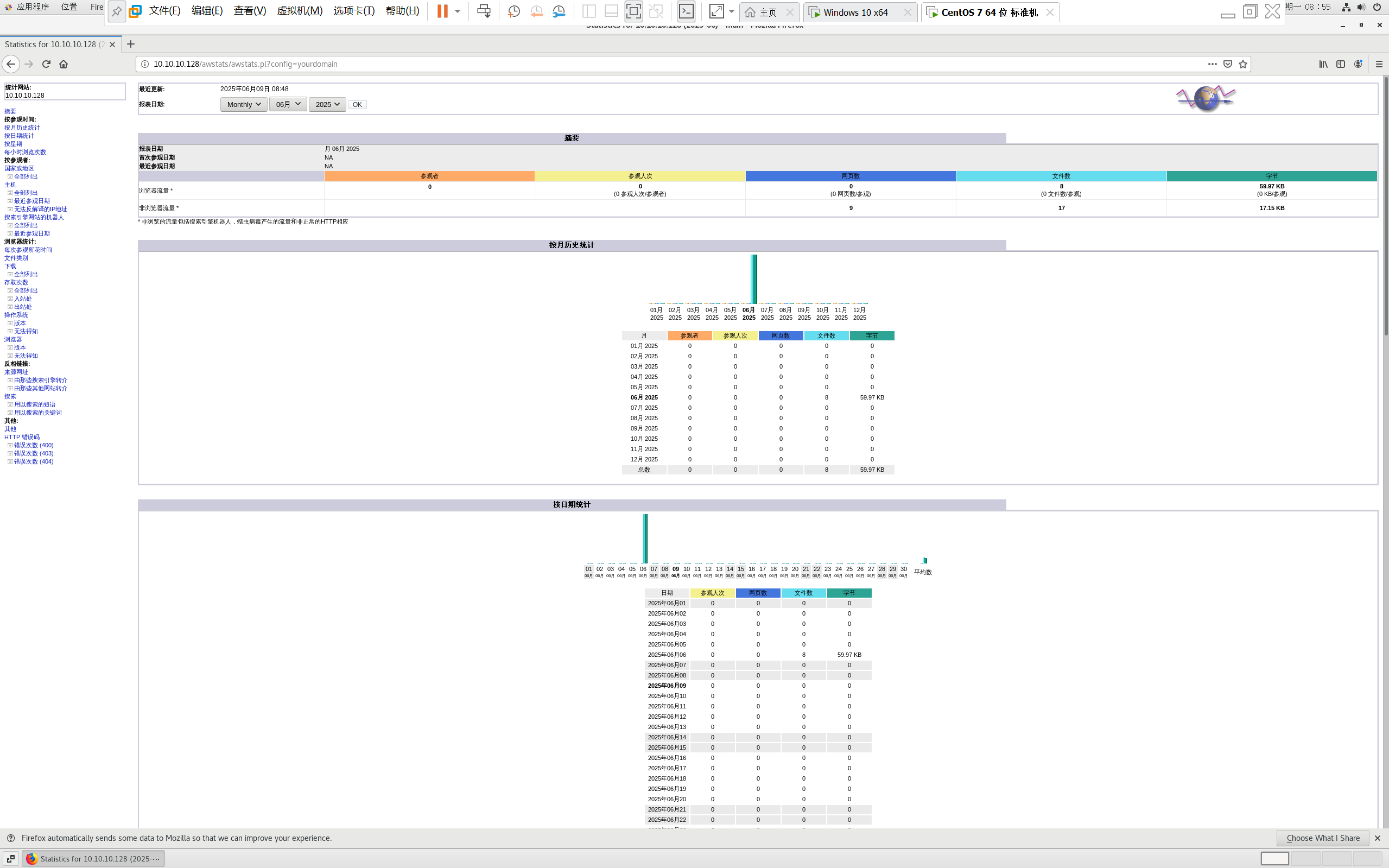
centos 7 部署awstats 网站访问检测
一、基础环境准备(两种安装方式都要做) bash # 安装必要依赖 yum install -y httpd perl mod_perl perl-Time-HiRes perl-DateTime systemctl enable httpd # 设置 Apache 开机自启 systemctl start httpd # 启动 Apache二、安装 AWStats࿰…...
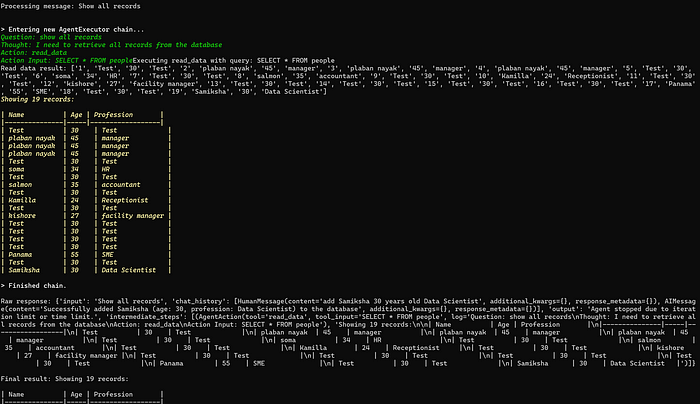
理解 MCP 工作流:使用 Ollama 和 LangChain 构建本地 MCP 客户端
🌟 什么是 MCP? 模型控制协议 (MCP) 是一种创新的协议,旨在无缝连接 AI 模型与应用程序。 MCP 是一个开源协议,它标准化了我们的 LLM 应用程序连接所需工具和数据源并与之协作的方式。 可以把它想象成你的 AI 模型 和想要使用它…...
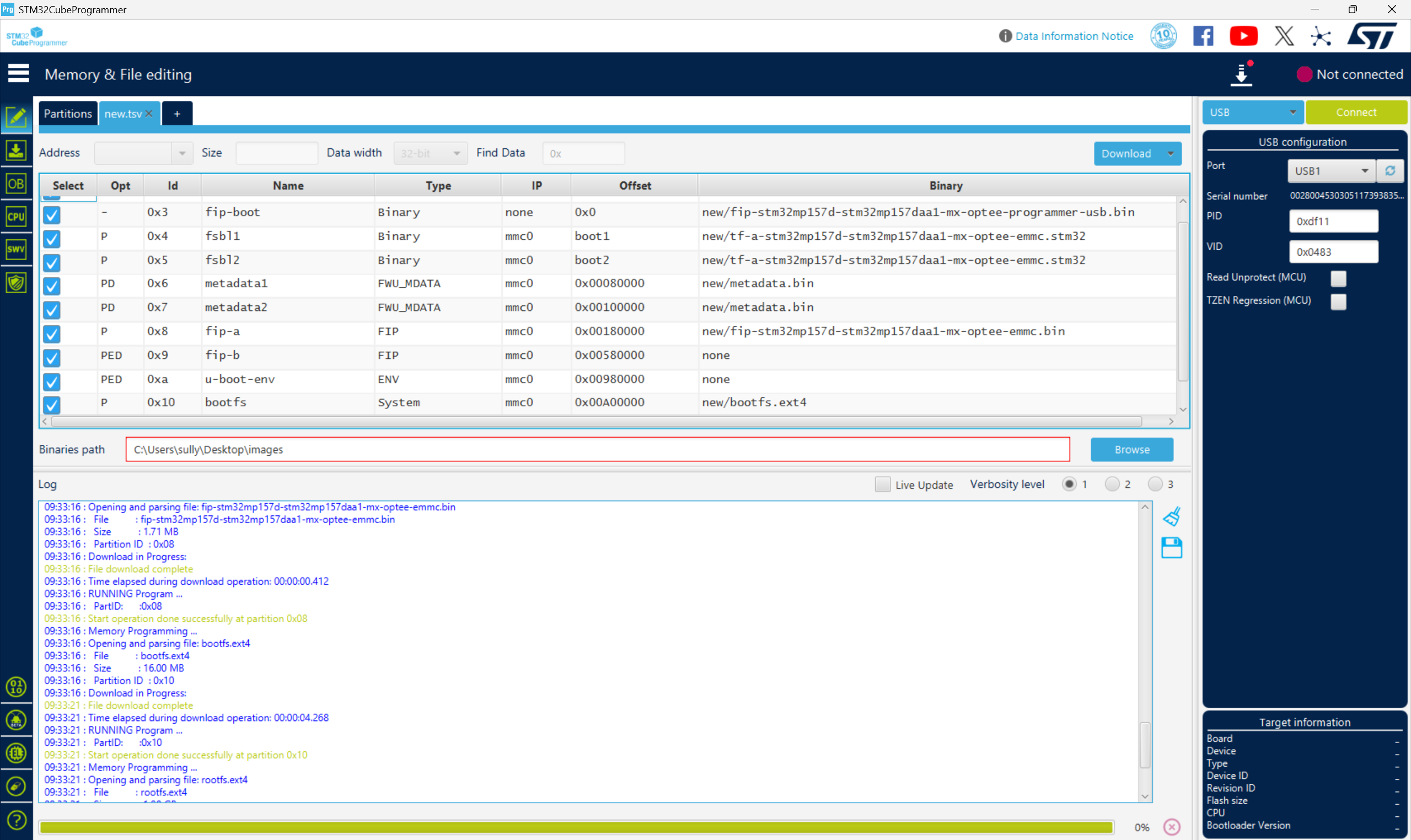
从零开始打造 OpenSTLinux 6.6 Yocto 系统(基于STM32CubeMX)(九)
设备树移植 和uboot设备树修改的内容同步到kernel将设备树stm32mp157d-stm32mp157daa1-mx.dts复制到内核源码目录下 源码修改及编译 修改arch/arm/boot/dts/st/Makefile,新增设备树编译 stm32mp157f-ev1-m4-examples.dtb \stm32mp157d-stm32mp157daa1-mx.dtb修改…...
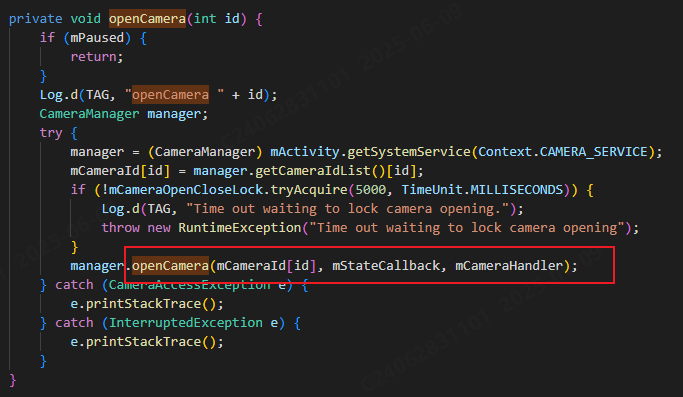
相机从app启动流程
一、流程框架图 二、具体流程分析 1、得到cameralist和对应的静态信息 目录如下: 重点代码分析: 启动相机前,先要通过getCameraIdList获取camera的个数以及id,然后可以通过getCameraCharacteristics获取对应id camera的capabilities(静态信息)进行一些openCamera前的…...

MySQL账号权限管理指南:安全创建账户与精细授权技巧
在MySQL数据库管理中,合理创建用户账号并分配精确权限是保障数据安全的核心环节。直接使用root账号进行所有操作不仅危险且难以审计操作行为。今天我们来全面解析MySQL账号创建与权限分配的专业方法。 一、为何需要创建独立账号? 最小权限原则…...

Java数值运算常见陷阱与规避方法
整数除法中的舍入问题 问题现象 当开发者预期进行浮点除法却误用整数除法时,会出现小数部分被截断的情况。典型错误模式如下: void process(int value) {double half = value / 2; // 整数除法导致截断// 使用half变量 }此时...
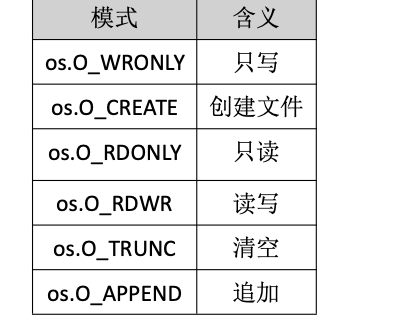
Golang——9、反射和文件操作
反射和文件操作 1、反射1.1、reflect.TypeOf()获取任意值的类型对象1.2、reflect.ValueOf()1.3、结构体反射 2、文件操作2.1、os.Open()打开文件2.2、方式一:使用Read()读取文件2.3、方式二:bufio读取文件2.4、方式三:os.ReadFile读取2.5、写…...
