其他:Python语言绘图合集




文章目录
- 介绍
- 注意
- 导入数据
- 函数模块
- 画图
介绍
python语言的科研绘图合集
注意
This dataset includes the following (All files are preceded by "Marle_et_al_Nature_AirborneFraction_"):- "Datasheet.xlsx": Excel dataset containing all annual and monthly emissions and CO2 time series used for the analysis, and the resulting airborne fraction time series.- "Script.py":
BEFORE RUNNING THE SCRIPT: change the 'wdir' variable to the directory containing the provided script and files.
NOTE: This script requires the Python module: 'pymannkendall'
Python script used for reproducing the results and figures from the paper. The provided Datasheet.xlsx file and the .zip and .npz files are required for this program. In case all these files are found by the script, it should run within several seconds. Successful execution of the script will save Figures 1-4 from the main text and print the data from Table 1. In case script execution takes longer, please check if the .xlsx, .zip and .npz files are correctly present in the assigned 'wdir' directory. Otherwise the script will start recalculating these files, which might take a while (see notes below).- "MC10000_MK_ts_TRENDabs.zip": .zip file containing all results from the Monte-Carlo simulation for trend estimation for Figure 3 (calculated using Python function 'calc_AF_MonteCarlo()'). This .zip file contains multiple .npz files for different emission scenarios and data treatments. This .zip file is managed by the Python script function 'calc_AF_MonteCarlo_filemanager()', there is no need to unzip the file manually. In case the .zip file is not found by the Python script (e.g. because the .zip file was unpacked manually and deleted), the program will start recalculating and save a new .zip file. This can take several minutes dependent on the computer used. Recalculated results could differ very slightly due to the random factor in the Monte-Carlo approach, even though the 10,000 iterations bring this variation to a minimum.- "MC1000_MK_run50x50_TRENDabs.npz": .npz file containing the Monte-Carlo results used for producing Figure 4 (calculated using Python function 'calc_AF_MonteCarlo_ARR()'). In case the .npz file is not found by the Python script (e.g. because it was deleted or not downloaded), the program will start recalculating and save a new file. This can take around 30 hours(!) dependent on the computer used. Recalculated results could differ slightly due to the random factor in the Monte-Carlo approach.- "tol_colors.py": Additional Python module used in script.py, required for producing the colors used in the Main text figures. Source: https://personal.sron.nl/~pault/- Figure files: Figures 1-4 from the Main text saved as .pdf files. Figure 3 is saved as three independent panels. The Figures are also reproduced by script.py if executed successfully.导入数据
数据可从以下链接下载(画图所需要的所有数据):
-
百度云盘链接: https://pan.baidu.com/s/1D9qBDOIxDRGtuvUMVKtxBA
-
提取码: fhp3
函数模块
讲下面代码存成tol_colors.py
import numpy as np
from matplotlib.colors import LinearSegmentedColormap, to_rgba_arraydef discretemap(colormap, hexclrs):"""Produce a colormap from a list of discrete colors without interpolation."""clrs = to_rgba_array(hexclrs)clrs = np.vstack([clrs[0], clrs, clrs[-1]])cdict = {}for ki, key in enumerate(('red','green','blue')):cdict[key] = [ (i/(len(clrs)-2.), clrs[i, ki], clrs[i+1, ki]) for i in range(len(clrs)-1) ]return LinearSegmentedColormap(colormap, cdict)class TOLcmaps(object):"""Class TOLcmaps definition."""def __init__(self):""""""self.cmap = Noneself.cname = Noneself.namelist = ('sunset_discrete', 'sunset', 'BuRd_discrete', 'BuRd','PRGn_discrete', 'PRGn', 'YlOrBr_discrete', 'YlOrBr', 'WhOrBr','iridescent', 'rainbow_PuRd', 'rainbow_PuBr', 'rainbow_WhRd','rainbow_WhBr', 'rainbow_discrete')self.funcdict = dict(zip(self.namelist,(self.__sunset_discrete, self.__sunset, self.__BuRd_discrete,self.__BuRd, self.__PRGn_discrete, self.__PRGn,self.__YlOrBr_discrete, self.__YlOrBr, self.__WhOrBr,self.__iridescent, self.__rainbow_PuRd, self.__rainbow_PuBr,self.__rainbow_WhRd, self.__rainbow_WhBr,self.__rainbow_discrete)))def __sunset_discrete(self):"""Define colormap 'sunset_discrete'."""clrs = ['#364B9A', '#4A7BB7', '#6EA6CD', '#98CAE1', '#C2E4EF','#EAECCC', '#FEDA8B', '#FDB366', '#F67E4B', '#DD3D2D','#A50026']self.cmap = discretemap(self.cname, clrs)self.cmap.set_bad('#FFFFFF')def __sunset(self):"""Define colormap 'sunset'."""clrs = ['#364B9A', '#4A7BB7', '#6EA6CD', '#98CAE1', '#C2E4EF','#EAECCC', '#FEDA8B', '#FDB366', '#F67E4B', '#DD3D2D','#A50026']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#FFFFFF')def __BuRd_discrete(self):"""Define colormap 'BuRd_discrete'."""clrs = ['#2166AC', '#4393C3', '#92C5DE', '#D1E5F0', '#F7F7F7','#FDDBC7', '#F4A582', '#D6604D', '#B2182B']self.cmap = discretemap(self.cname, clrs)self.cmap.set_bad('#FFEE99')def __BuRd(self):"""Define colormap 'BuRd'."""clrs = ['#2166AC', '#4393C3', '#92C5DE', '#D1E5F0', '#F7F7F7','#FDDBC7', '#F4A582', '#D6604D', '#B2182B']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#FFEE99')def __PRGn_discrete(self):"""Define colormap 'PRGn_discrete'."""clrs = ['#762A83', '#9970AB', '#C2A5CF', '#E7D4E8', '#F7F7F7','#D9F0D3', '#ACD39E', '#5AAE61', '#1B7837']self.cmap = discretemap(self.cname, clrs)self.cmap.set_bad('#FFEE99')def __PRGn(self):"""Define colormap 'PRGn'."""clrs = ['#762A83', '#9970AB', '#C2A5CF', '#E7D4E8', '#F7F7F7','#D9F0D3', '#ACD39E', '#5AAE61', '#1B7837']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#FFEE99')def __YlOrBr_discrete(self):"""Define colormap 'YlOrBr_discrete'."""clrs = ['#FFFFE5', '#FFF7BC', '#FEE391', '#FEC44F', '#FB9A29','#EC7014', '#CC4C02', '#993404', '#662506']self.cmap = discretemap(self.cname, clrs)self.cmap.set_bad('#888888')def __YlOrBr(self):"""Define colormap 'YlOrBr'."""clrs = ['#FFFFE5', '#FFF7BC', '#FEE391', '#FEC44F', '#FB9A29','#EC7014', '#CC4C02', '#993404', '#662506']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#888888')def __WhOrBr(self):"""Define colormap 'WhOrBr'."""clrs = ['#FFFFFF', '#FFF7BC', '#FEE391', '#FEC44F', '#FB9A29','#EC7014', '#CC4C02', '#993404', '#662506']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#888888')def __iridescent(self):"""Define colormap 'iridescent'."""clrs = ['#FEFBE9', '#FCF7D5', '#F5F3C1', '#EAF0B5', '#DDECBF','#D0E7CA', '#C2E3D2', '#B5DDD8', '#A8D8DC', '#9BD2E1','#8DCBE4', '#81C4E7', '#7BBCE7', '#7EB2E4', '#88A5DD','#9398D2', '#9B8AC4', '#9D7DB2', '#9A709E', '#906388','#805770', '#684957', '#46353A']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#999999')def __rainbow_PuRd(self):"""Define colormap 'rainbow_PuRd'."""clrs = ['#6F4C9B', '#6059A9', '#5568B8', '#4E79C5', '#4D8AC6','#4E96BC', '#549EB3', '#59A5A9', '#60AB9E', '#69B190','#77B77D', '#8CBC68', '#A6BE54', '#BEBC48', '#D1B541','#DDAA3C', '#E49C39', '#E78C35', '#E67932', '#E4632D','#DF4828', '#DA2222']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#FFFFFF')def __rainbow_PuBr(self):"""Define colormap 'rainbow_PuBr'."""clrs = ['#6F4C9B', '#6059A9', '#5568B8', '#4E79C5', '#4D8AC6','#4E96BC', '#549EB3', '#59A5A9', '#60AB9E', '#69B190','#77B77D', '#8CBC68', '#A6BE54', '#BEBC48', '#D1B541','#DDAA3C', '#E49C39', '#E78C35', '#E67932', '#E4632D','#DF4828', '#DA2222', '#B8221E', '#95211B', '#721E17','#521A13']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#FFFFFF')def __rainbow_WhRd(self):"""Define colormap 'rainbow_WhRd'."""clrs = ['#E8ECFB', '#DDD8EF', '#D1C1E1', '#C3A8D1', '#B58FC2','#A778B4', '#9B62A7', '#8C4E99', '#6F4C9B', '#6059A9','#5568B8', '#4E79C5', '#4D8AC6', '#4E96BC', '#549EB3','#59A5A9', '#60AB9E', '#69B190', '#77B77D', '#8CBC68','#A6BE54', '#BEBC48', '#D1B541', '#DDAA3C', '#E49C39','#E78C35', '#E67932', '#E4632D', '#DF4828', '#DA2222']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#666666')def __rainbow_WhBr(self):"""Define colormap 'rainbow_WhBr'."""clrs = ['#E8ECFB', '#DDD8EF', '#D1C1E1', '#C3A8D1', '#B58FC2','#A778B4', '#9B62A7', '#8C4E99', '#6F4C9B', '#6059A9','#5568B8', '#4E79C5', '#4D8AC6', '#4E96BC', '#549EB3','#59A5A9', '#60AB9E', '#69B190', '#77B77D', '#8CBC68','#A6BE54', '#BEBC48', '#D1B541', '#DDAA3C', '#E49C39','#E78C35', '#E67932', '#E4632D', '#DF4828', '#DA2222','#B8221E', '#95211B', '#721E17', '#521A13']self.cmap = LinearSegmentedColormap.from_list(self.cname, clrs)self.cmap.set_bad('#666666')def __rainbow_discrete(self, lut=None):"""Define colormap 'rainbow_discrete'."""clrs = ['#E8ECFB', '#D9CCE3', '#D1BBD7', '#CAACCB', '#BA8DB4','#AE76A3', '#AA6F9E', '#994F88', '#882E72', '#1965B0','#437DBF', '#5289C7', '#6195CF', '#7BAFDE', '#4EB265','#90C987', '#CAE0AB', '#F7F056', '#F7CB45', '#F6C141','#F4A736', '#F1932D', '#EE8026', '#E8601C', '#E65518','#DC050C', '#A5170E', '#72190E', '#42150A']indexes = [[9], [9, 25], [9, 17, 25], [9, 14, 17, 25], [9, 13, 14, 17,25], [9, 13, 14, 16, 17, 25], [8, 9, 13, 14, 16, 17, 25], [8,9, 13, 14, 16, 17, 22, 25], [8, 9, 13, 14, 16, 17, 22, 25, 27],[8, 9, 13, 14, 16, 17, 20, 23, 25, 27], [8, 9, 11, 13, 14, 16,17, 20, 23, 25, 27], [2, 5, 8, 9, 11, 13, 14, 16, 17, 20, 23,25], [2, 5, 8, 9, 11, 13, 14, 15, 16, 17, 20, 23, 25], [2, 5,8, 9, 11, 13, 14, 15, 16, 17, 19, 21, 23, 25], [2, 5, 8, 9, 11,13, 14, 15, 16, 17, 19, 21, 23, 25, 27], [2, 4, 6, 8, 9, 11,13, 14, 15, 16, 17, 19, 21, 23, 25, 27], [2, 4, 6, 7, 8, 9, 11,13, 14, 15, 16, 17, 19, 21, 23, 25, 27], [2, 4, 6, 7, 8, 9, 11,13, 14, 15, 16, 17, 19, 21, 23, 25, 26, 27], [1, 3, 4, 6, 7, 8,9, 11, 13, 14, 15, 16, 17, 19, 21, 23, 25, 26, 27], [1, 3, 4,6, 7, 8, 9, 10, 12, 13, 14, 15, 16, 17, 19, 21, 23, 25, 26,27], [1, 3, 4, 6, 7, 8, 9, 10, 12, 13, 14, 15, 16, 17, 18, 20,22, 24, 25, 26, 27], [1, 3, 4, 6, 7, 8, 9, 10, 12, 13, 14, 15,16, 17, 18, 20, 22, 24, 25, 26, 27, 28], [0, 1, 3, 4, 6, 7, 8,9, 10, 12, 13, 14, 15, 16, 17, 18, 20, 22, 24, 25, 26, 27, 28]]if lut == None or lut < 1 or lut > 23:lut = 22self.cmap = discretemap(self.cname, [ clrs[i] for i in indexes[lut-1] ])if lut == 23:self.cmap.set_bad('#777777')else:self.cmap.set_bad('#FFFFFF')def show(self):"""List names of defined colormaps."""print(' '.join(repr(n) for n in self.namelist))def get(self, cname='rainbow_PuRd', lut=None):"""Return requested colormap, default is 'rainbow_PuRd'."""self.cname = cnameif cname == 'rainbow_discrete':self.__rainbow_discrete(lut)else:self.funcdict[cname]()return self.cmapdef tol_cmap(colormap=None, lut=None):"""Continuous and discrete color sets for ordered data.Return a matplotlib colormap.Parameter lut is ignored for all colormaps except 'rainbow_discrete'."""obj = TOLcmaps()if colormap == None:return obj.namelistif colormap not in obj.namelist:colormap = 'rainbow_PuRd'print('*** Warning: requested colormap not defined,','known colormaps are {}.'.format(obj.namelist),'Using {}.'.format(colormap))return obj.get(colormap, lut)def tol_cset(colorset=None):"""Discrete color sets for qualitative data.Define a namedtuple instance with the colors.Examples for: cset = tol_cset(<scheme>)- cset.red and cset[1] give the same color (in default 'bright' colorset)- cset._fields gives a tuple with all color names- list(cset) gives a list with all colors"""from collections import namedtuplenamelist = ('bright', 'high-contrast', 'vibrant', 'muted', 'medium-contrast', 'light')if colorset == None:return namelistif colorset not in namelist:colorset = 'bright'print('*** Warning: requested colorset not defined,','known colorsets are {}.'.format(namelist),'Using {}.'.format(colorset)) if colorset == 'bright':cset = namedtuple('Bcset','blue red green yellow cyan purple grey black')return cset('#4477AA', '#EE6677', '#228833', '#CCBB44', '#66CCEE','#AA3377', '#BBBBBB', '#000000')if colorset == 'high-contrast':cset = namedtuple('Hcset','blue yellow red black')return cset('#004488', '#DDAA33', '#BB5566', '#000000')if colorset == 'vibrant':cset = namedtuple('Vcset','orange blue cyan magenta red teal grey black')return cset('#EE7733', '#0077BB', '#33BBEE', '#EE3377', '#CC3311','#009988', '#BBBBBB', '#000000')if colorset == 'muted':cset = namedtuple('Mcset','rose indigo sand green cyan wine teal olive purple pale_grey black')return cset('#CC6677', '#332288', '#DDCC77', '#117733', '#88CCEE','#882255', '#44AA99', '#999933', '#AA4499', '#DDDDDD','#000000')if colorset == 'medium-contrast':cset = namedtuple('Mcset','light_blue dark_blue light_yellow dark_red dark_yellow light_red black')return cset('#6699CC', '#004488', '#EECC66', '#994455', '#997700','#EE99AA', '#000000')if colorset == 'light':cset = namedtuple('Lcset','light_blue orange light_yellow pink light_cyan mint pear olive pale_grey black')return cset('#77AADD', '#EE8866', '#EEDD88', '#FFAABB', '#99DDFF','#44BB99', '#BBCC33', '#AAAA00', '#DDDDDD', '#000000')def main():from matplotlib import pyplot as plt# Change default colorset (for lines) and colormap (for maps).
# plt.rc('axes', prop_cycle=plt.cycler('color', list(tol_cset('bright'))))
# plt.cm.register_cmap('rainbow_PuRd', tol_cmap('rainbow_PuRd'))
# plt.rc('image', cmap='rainbow_PuRd')# Show colorsets tol_cset(<scheme>).schemes = tol_cset()fig, axes = plt.subplots(ncols=len(schemes), figsize=(9, 3))fig.subplots_adjust(top=0.9, bottom=0.02, left=0.02, right=0.92)for ax, scheme in zip(axes, schemes):cset = tol_cset(scheme)names = cset._fieldscolors = list(cset)for name, color in zip(names, colors):ax.scatter([], [], c=color, s=80, label=name)ax.set_axis_off()ax.legend(loc=2)ax.set_title(scheme)plt.show()# Show colormaps tol_cmap(<scheme>). schemes = tol_cmap()gradient = np.linspace(0, 1, 256)gradient = np.vstack((gradient, gradient))fig, axes = plt.subplots(nrows=len(schemes))fig.subplots_adjust(top=0.98, bottom=0.02, left=0.2, right=0.99)for ax, scheme in zip(axes, schemes):pos = list(ax.get_position().bounds)ax.set_axis_off()ax.imshow(gradient, aspect=4, cmap=tol_cmap(scheme))fig.text(pos[0] - 0.01, pos[1] + pos[3]/2., scheme, va='center', ha='right', fontsize=10)plt.show()# Show colormaps tol_cmap('rainbow_discrete', <lut>). gradient = np.linspace(0, 1, 256)gradient = np.vstack((gradient, gradient))fig, axes = plt.subplots(nrows=23)fig.subplots_adjust(top=0.98, bottom=0.02, left=0.25, right=0.99)for lut, ax in enumerate(axes, start=1):pos = list(ax.get_position().bounds)ax.set_axis_off()ax.imshow(gradient, aspect=4, cmap=tol_cmap('rainbow_discrete', lut))fig.text(pos[0] - 0.01, pos[1] + pos[3]/2., 'rainbow_discrete, ' + str(lut), va='center', ha='right', fontsize=10)plt.show()if __name__ == '__main__':main()
画图
# Python standard library
import os
import io
import sys
import time as timer
from zipfile import ZipFile
from collections import OrderedDict
# Scientific modules
import numpy as np
import pandas as pd
import scipy.optimize
import scipy.stats
import scipy.ndimage
import pymannkendall as mk
import matplotlib.pyplot as plt
import matplotlib.colors as pltc
import matplotlib as mpl
from matplotlib.ticker import MultipleLocator
import matplotlib# Change standard values
plt.rc('image', interpolation='none') # Change default image interpolation
mm = 1/25.4 # Conversion factor inches to mm for figure sizes.# Set working directory
wdir = '/Path/to/directory/' # Change to directory containing this script and the data.sys.path.append(wdir)# Additional modules provided with this script
import tol_colors# Define functions
def anom(x, order=2):''' Split time series into anomaly and trend componentscalculation of anomaly by removal of 2-order polynomial trend and climatology:param x: time series [1D array]:param order: order of polynomial fit [int] (default=2):return: x_A (anomaly component), x_T (trend component)'''t_m = range(len(x))fitx = np.polyfit(t_m, x, order)fit_fn = np.poly1d(fitx) # fit functionx_T = fit_fn(t_m) # x_T is trend componentx_F = x - x_T # removal of trendx_C = np.mean(np.reshape(x_F, (int(len(t_m) / 12.), 12)), 0) # x_C is climatologyx_A = x_F - np.tile(x_C, int(len(t_m) / 12.)) # x_A is anomaly, removal of climatologyreturn x_A, x_Tdef norm(x):''' Normalize time series:param x: time series [1D array]:return: normalized time series'''return (x - np.mean(x)) / np.std(x)def tomonthly(x):''' Interpolate annual flux to monthly flux:param x: annual time series [1D array]:return: interpolated monthly time series'''return np.interp(np.linspace(0, len(x)-1/12., len(x)*12), np.linspace(0, len(x)-1, len(x)), x) / 12.def amean(x):''' Calculate annual average:param x: monthly time series [1D array]:return: annual mean'''if len(x) % 12 == 0:return np.nanmean(np.reshape(x, (int(len(x) / 12.), 12)), 1) # annual averageelse:return xdef lambdafn(x_A, ENSOI_m, VOLC_m, inverse=True):''' Function for determination of lambda:param x_A: time series anomaly [1D array]:param ENSOI_m: monthly ENSO index [1D array]:param VOLC_m: monthly volcanic index [1D array]:param inverse: minimize based on the inverse correlation coefficient [bool] (default=True):return: lambda and corresponding maximum correlation'''if inverse == True:def fuevi(lam): return 1-np.corrcoef((ENSOI_m + lam * VOLC_m, x_A))[0,1]elif inverse == False:def fuevi(lam): return np.corrcoef((ENSOI_m + lam * VOLC_m, x_A))[0,1]lam_maxcor = scipy.optimize.minimize(fuevi, x0=0.5)['x'][0] # maximize correlation between EVI and x_Amaxcor = fuevi(lam_maxcor)return lam_maxcor, maxcordef filterfn(x_A, x_T, ref):'''Filtering of timeseries by ENSO or EVI index.:param x_A: time series anomaly component [1D array]:param x_T: time series trend component [1D array]:param ref: reference index = ENSOI or EVI [1D array]:return: filtered time series and corresponding mu of minimum variance'''def fu2(mu): return np.var(x_A - mu*ref)mu_minvar = scipy.optimize.minimize(fu2, x0=1)['x'][0] # determine mu by minimization of variance.x_U = x_A - mu_minvar*refx_n = x_T + x_U # x_n is the filtered anomalyreturn x_n, mu_minvardef rgba2rgb(rgba, background=None):''' Convert rgba values to rgb. Source: https://stackoverflow.com/questions/50331463/convert-rgba-to-rgb-in-python:param rgba: rgba values [array]:param background: background rgb value [tuple] (default = (255,255,255)):return: rgb values'''if background is None:background = (255, 255, 255)R, G, B = backgroundif rgba.ndim == 3: row, col, ch = rgba.shape # 2d array of colors.elif rgba.ndim == 2: row, ch = rgba.shape # 1d array of colors.elif rgba.ndim == 1: ch, = rgba.shapeif ch == 3:return rgbaassert ch == 4, 'RGBA image has 4 channels.'if rgba.ndim == 3:rgb = np.zeros((row, col, 3), dtype='float32')r, g, b, a = rgba[:, :, 0], rgba[:, :, 1], rgba[:, :, 2], rgba[:, :, 3]a = np.asarray(a, dtype='float32') / 255.rgb[:, :, 0] = r * a + (1.0 - a) * Rrgb[:, :, 1] = g * a + (1.0 - a) * Grgb[:, :, 2] = b * a + (1.0 - a) * Belif rgba.ndim == 2:rgb = np.zeros((row, 3), dtype='float32')r, g, b, a = rgba[:, 0], rgba[:, 1], rgba[:, 2], rgba[:, 3]a = np.asarray(a, dtype='float32') / 255.rgb[:, 0] = r * a + (1.0 - a) * Rrgb[:, 1] = g * a + (1.0 - a) * Grgb[:, 2] = b * a + (1.0 - a) * Belif rgba.ndim == 1:rgb = np.zeros((3), dtype='float32')r, g, b, a = rgba[0], rgba[1], rgba[2], rgba[3]a = np.asarray(a, dtype='float32') / 255.rgb[0] = r * a + (1.0 - a) * Rrgb[1] = g * a + (1.0 - a) * Grgb[2] = b * a + (1.0 - a) * Breturn np.asarray(rgb, dtype='uint8')def mktest(t, y):''' Perform Mann-Kendall test for trend estimation.:param t: time axis [1D array]:param y: data time series [1D array]:return: Mann-Kendall test results'''mk_test = mk.original_test(y)# Intercept has to extrapolated to x=0, in order to get similar output format as scipy.stats.linregress.return (mk_test.slope, mk_test.intercept - mk_test.slope * t[0], 0, mk_test.p, 0) # placed in tuple to get same output format as scipy.linregress.def slopest(t, y, method):''' Wrapper for choosing trend estimation based on linear regression or Mann-Kendall test:param t: time axis [1D array]:param y: data time series [1D array]:param method: linear regression ('linregres') or Mann-Kendall test ('mannkendall') [str]:return: Regression results'''if method == 'linregres':return scipy.stats.linregress(t, y)elif method == 'mannkendall':return mktest(t, y)def mvavg(x, mavg, edge=False):''' Calculate moving average:param x: monthly or annual time series [1D array]:param mavg: window size [int]:param edge: choose how to deal with time series edges [bool] (default = False):return: y, averaged time series'''if edge is True:if mavg % 2 == 0: # if number is even.x = np.hstack((np.tile(np.mean(x[:int(mavg/2.)]),int(mavg/2.)), x, np.tile(np.mean(x[int(np.floor(-mavg/2.)):]),int(mavg/2.)-1))) else: # if number is odd.x = np.hstack((np.tile(np.mean(x[:int(mavg/2.)]),int(mavg/2.)), x, np.tile(np.mean(x[int(np.floor(-mavg/2.)):]),int(mavg/2.))))y = np.convolve(x, np.ones(mavg) / float(mavg), 'valid')else:y = np.convolve(x, np.ones(mavg) / float(mavg), 'same')return ydef load_colors_for_plots():''' Loads line colors, categorial colors and colormap, for use in plotting. Source: https://personal.sron.nl/~pault/:return: colors, ccat_fig1, ccat_fig2, cmap'''# Load line colors for all Figurescolors = tol_colors.tol_cset('high-contrast')colors = [colors.red, colors.yellow, colors.blue]# Load category colors for Figure 1.ccat_fig1 = tol_colors.tol_cset('bright')ccat_fig1b = tol_colors.tol_cset('high-contrast')ccat_fig1 = [ccat_fig1.yellow, ccat_fig1b.red, ccat_fig1.green]# Load category colors for Figure 2.ccat_fig2 = tol_colors.tol_cset('bright')ccat_fig2 = [ccat_fig2.yellow, ccat_fig2.grey, 'black', ccat_fig2.red, ccat_fig2.purple, ccat_fig2.cyan, ccat_fig2.green]# Load colormap for Figure 4.cmap = plt.get_cmap('RdBu_r', 100)return colors, ccat_fig1, ccat_fig2, cmapdef data_selector(lustr, FF_China=None, RESPscheme=None):''' Choose to load one of the three land use emissions dataset time series:param lustr: 'gcp' (Global Carbon Project), 'han' (Houghton & Nassikas), or 'new' (This study):param FF_China: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param RESPscheme: choose respiration scheme, normal ('stable') or increasing role of fire ('incfire'):return: tuples with LULCC and FF emission time series.'''if FF_China is None: FF_China = 'GCP'if RESPscheme is None: RESPscheme = 'stable'if lustr == 'gcp':lu_tuple = (LUgcp, LUgcp_m, LUgcp_error, LUgcp_error_m)elif lustr == 'han':lu_tuple = (LUhan, LUhan_m, LUhan_error, LUhan_error_m)elif lustr == 'new':if RESPscheme == 'stable':lu_tuple = (LUnew, LUnew_m, LUnew_error, LUnew_error_m)elif RESPscheme == 'incfire':lu_tuple = (datadict['Total_incfire'], datadict['Total_incfire_m'], LUnew_error, LUnew_error_m)# Select used fossil fuel time seriesif FF_China == 'GCP':ff_tuple = (FFgcp, FFgcp_m, FFgcp_error, FFgcp_error_m)elif FF_China == 'LIU':ff_tuple = (FFadj, FFadj_m, FFadj_error, FFadj_error_m)elif FF_China == 'BP':ff_tuple = (FFadjBP, FFadjBP_m, FFadjBP_error, FFadjBP_error_m)return lu_tuple, ff_tuple# Load data from Excel file
emissions = pd.read_excel(wdir + 'Marle_et_al_Nature_AirborneFraction_Datasheet.xlsx', 'Emissions')
atmosphere = pd.read_excel(wdir + 'Marle_et_al_Nature_AirborneFraction_Datasheet.xlsx', 'Atmosphere')
atmosphere_m = pd.read_excel(wdir + 'Marle_et_al_Nature_AirborneFraction_Datasheet.xlsx', 'Atmosphere_monthly')
faostat = pd.read_excel(wdir + 'Marle_et_al_Nature_AirborneFraction_Datasheet.xlsx', 'FAOStat', index_col=0, header=6) # Load soy, palmoil data# Settings
C2CO2 = 3.66 # C to C02 conversion factor.
fPG = 2.134 # factor ppm CO2 to Pg C.year0 = 1959 # default = 1959
year1 = 2019 # defualt = 2019
years = np.arange(year0, year1+1) # annual time vector
t_y = np.arange(len(years)) + int(years[0]) # annual time vector for plots
t_m = np.arange(0.0, len(years), 1 / 12.0) + int(years[0]) # monthly time vector for plotsmavg = 15 # Moving average window (default = 15)
edge = int(np.floor(mavg/2.0)) # extra edge values required for moving averageiset = 3 # Which AF time series to use for plotting (default=3). 0 = annual raw ('AF_a'), 1 = monthly smoothed ('AF_ms'), 2 = monthly smoothed and filtered ('AF_msnv'), 3 = annual smoothed and filtered ('AF_asnv')trendcalc = 'abs' # absolute trend per decade ('abs', default), or relative growth rate ('rgr'; not fully tested, could give errors)
trendmethod = 'mannkendall' # Linear regression ('linregres') or Mann-Kendall test ('mannkendall', default)# unit conversion factors
if trendcalc == 'abs': uconv = 10 # per year to per decade.
elif trendcalc == 'rgr': uconv = 100 # fraction to percentage.
cconv = 0.001 # conversion from Tg C to Pg C.# Load colorsets for plots
colors, ccat_fig1, ccat_fig2, cmap = load_colors_for_plots()
Cgcp, Chan, Cnew = colors# find index of start year and end year in data sheets
index0 = np.where(emissions['A'] == year0)[0][0] # annual indices
index1 = np.where(emissions['A'] == year1)[0][0]
index0_m = np.where(atmosphere_m['A'] == year0)[0][0] # monthly indices
index1_m = np.where(atmosphere_m['A'] == year1)[0][0]+12-1# Load fossil fuel emissions
FFgcp = np.array(emissions['N'][index0:index1+1]).astype(float) * cconv
FFchinaGCP = np.array(emissions['O'][index0:index1+1]).astype(float) * cconv
FFchinaLIU = np.array(emissions['P'][index0:index1+1]).astype(float) * cconv
FFchinaBP = np.array(emissions['Q'][index0:index1+1]).astype(float) * cconv
FFadj = FFgcp - FFchinaGCP + FFchinaLIU # Fossil fuels with China emissions replaced by Liu et al. (2015)
FFadjBP = FFgcp - FFchinaGCP + FFchinaBP # Fossil fuels with China emissions replaced by BP# Load land-use change emissions
LUgcp = np.array(emissions['M'][index0:index1+1]).astype(float) * cconv # LULCC emissions from Global Carbon Project (2020)
LUhan = np.array(emissions['G'][index0:index1+1]).astype(float) * cconv # LULCC emissions from Houghton & Nassikas (2017)
LUnew = np.array(emissions['L'][index0:index1+1]).astype(float) * cconv # LULCC emissions from This Study.# Load atmospheric CO2
CO2mlo = atmosphere['B'][index0:index1+2] # Atmospheric CO2 Mauna Loa. index +2 because one extra value needed for np.diff.
CO2spo = atmosphere['C'][index0:index1+2] # Atmospheric CO2 South Pole.
CO2 = np.mean([CO2mlo, CO2spo], axis=0).astype(float) # Average of MLO and SPO
CO2 = (CO2 - 280) * fPG # Subtract pre-industrial 280 ppm, convert to Pg C.
CO2_m = np.mean([atmosphere_m['C'][index0_m:index1_m+2], atmosphere_m['D'][index0_m:index1_m+2]], axis=0).astype(float) # Monthly average of MLO and SPO. index +2 because one extra value needed for np.diff.
CO2_m = (CO2_m - 280) * fPG # Subtract pre-industrial 280 ppm, convert to Pg C.
dCO2 = np.diff(CO2)
dCO2_m = np.diff(CO2_m) # -11 instead of -12 because one extra month is needed for np.diff.# Load ENSO and volcanic indices
ENSO_m = np.array(atmosphere_m['E'][index0_m-12:index1_m+1]).astype(float) # load one year extra (1958), required for 4 month lag.
VOLC_m = np.array(atmosphere_m['F'][index0_m:index1_m+1]).astype(float)# Define annual errors
LUgcp_error = LUgcp * 0.5 # 50%. Corresponds to about 0.7 Pg C yr-1 in GCP (2020)
LUhan_error = LUhan * 0.5 # 50%
LUnew_error = LUnew * 0.5 # 50%
FFgcp_error = FFgcp * 0.05 # 5%
FFadj_error = (FFgcp - FFchinaGCP) * 0.05 + FFchinaLIU * 0.10 # GCP error=5%, Liu error=10%
FFadjBP_error = (FFgcp - FFchinaGCP) * 0.05 + FFchinaBP * 0.10 # GCP error=5%, BP error=10%
dCO2_error = np.array(atmosphere['E'][index0:index1+1]).astype(float) * fPG# Interpolate annual fluxes to monthly fluxes
LUgcp_m = tomonthly(LUgcp)
LUhan_m = tomonthly(LUhan)
LUnew_m = tomonthly(LUnew)
FFgcp_m = tomonthly(FFgcp)
FFadj_m = tomonthly(FFadj)
FFadjBP_m = tomonthly(FFadjBP)# Calculate monthly error
LUgcp_error_m = LUgcp_m * 0.5
LUhan_error_m = LUhan_m * 0.5
LUnew_error_m = LUnew_m * 0.5
FFgcp_error_m = FFgcp_m * 0.05
FFadj_error_m = ((FFgcp_m - tomonthly(FFchinaGCP)) * 0.05 + tomonthly(FFchinaLIU) * 0.10) # GCP error=5%, Liu error=10%
FFadjBP_error_m = ((FFgcp_m - tomonthly(FFchinaGCP)) * 0.05 + tomonthly(FFchinaBP) * 0.10) # GCP error=5%, BP error=10%
dCO2_error_m = np.repeat(dCO2_error, 12) / 12.# Calculate EVI
ENSOI_m = norm(ENSO_m[12-4:-4]) # effect from enso lagged by 4 months and normalized to get enso indexdCO2_A, dCO2_T = anom(dCO2_m) # Seperate anomaly and trend
dCO2_As = mvavg(dCO2_A, mavg) # Apply moving average to anomaly
dCO2_s = mvavg(dCO2_m, mavg)lam_maxcor_dCO2_As, maxcor_dCO2_As = lambdafn(dCO2_As, ENSOI_m, VOLC_m, inverse=True) # Calculate maximum correlation between enso and volcanic indices.
lam_maxcor = -16 # lambda ~= -16 (Raupach, 2008), i.e. volcanic contribution to ensoi
EVI_m = norm(ENSOI_m + lam_maxcor * VOLC_m) # Calculate index of combination of enso and volcanos, called EVI.
ENSOI_A, ENSOI_T = anom(ENSOI_m) # ensoi anomaly and trend
EVI_A, EVI_T = anom(EVI_m) # EVI anomaly and trendENSOI_s = norm(mvavg(ENSOI_m, mavg)) # moving average with time window mavg, and normalize again
ENSOI_As = norm(mvavg(ENSOI_A, mavg))
EVI_s = norm(mvavg(EVI_m, mavg))
EVI_As = norm(mvavg(EVI_A, mavg))# Filter atmospheric CO2 growth rate using EVI index
dCO2_sv_m, mu_minvar = filterfn(dCO2_As, dCO2_T, EVI_As/12)
dCO2_sv = np.sum(np.reshape(dCO2_sv_m, [len(years), 12]), 1) # annual sum# Set up respiration schemes
fraction_stable = np.ones(index1-index0+1) # stable fraction of fire emissions that is thought to respire (main case)
fraction_incfire = np.linspace(3, 1, year1-year0+1) # increasing fraction of fire emissions that is thought to respire (increasing role of fire) # Load individual LUnew emission components [labels, Excel column, plot color]
metadict = OrderedDict([('Other', ['Other regions', 'J', ccat_fig2[0]]),('EQAS_peatdrain', ['EQAS peat drainage', 'K', ccat_fig2[1]]),('EQAS_deco', ['EQAS decomposition', 'F', ccat_fig2[2]]),('EQAS_peatfire', ['EQAS peat fire', 'E', ccat_fig2[3]]),('EQAS_fire', ['EQAS fire', 'D', ccat_fig2[4]]),('ARCD_deco', ['ARCD decomposition', 'C', ccat_fig2[5]]),('ARCD_fire', ['ARCD fire', 'B', ccat_fig2[6]]),])datadict = OrderedDict()
for keyn in metadict.keys():data = np.array(emissions[metadict[keyn][1]][index0:index1 + 1]).astype(float)if keyn in ['ARCD_deco', 'EQAS_deco']: # For decomposition emissions calculate also calculate the 'increasing role of fire' scenario.datadict[keyn] = data * fraction_stable * cconvdatadict[keyn + '_incfire'] = data * fraction_incfire * cconvelse:datadict[keyn] = data * cconv
del datadatadict['Total'] = np.sum([datadict[keyn] for keyn in metadict.keys()], axis=0) # World LULCC emissions, identical to LUnew.
datadict['Total_incfire'] = datadict['Total'] - datadict['EQAS_deco'] - datadict['ARCD_deco'] + datadict['EQAS_deco_incfire'] + datadict['ARCD_deco_incfire'] # 'increasing role of fire' scenario.
datadict['Total_incfire_m'] = tomonthly(datadict['Total_incfire']) # interpolate to monthly time series.def plot_Figure1(filename=None):''' Plot paper Figure 1. Emissions distribution.Creates plot with stacked values of ARCD: Emissions + Decomposition + line with soy bean and cattle export,EQAS: Emissions + Peat Drainage + Decomposition + line with palm oil export:param fign: figure number:param filename: filename of saved figure (.pdf format). If filename == None, figure is only displayed and not saved.:return: Figure 1'''fontsize = 7fontsize_legend = 6.5fontsize_panellabel = 8align = 'center'plt.rcParams['hatch.linewidth'] = 0.3fig, (ax1,ax2) = plt.subplots(2, 1, figsize=(120*mm,80*mm))p1 = ax1.bar(t_y, datadict['ARCD_deco'], align=align, color=ccat_fig1[0], edgecolor='white', linewidth=0.6, label='Decomposition') # , hatch='')p2 = ax1.bar(t_y, datadict['ARCD_fire'], bottom=datadict['ARCD_deco'], align=align, color=ccat_fig1[1], edgecolor='white', linewidth=0.6, label='Fire emissions') # , hatch='\\\\\\\\')# Add soybean and cattle exportax11 = ax1.twinx() l1 = ax11.plot(t_y[2:], (faostat['ARCD_SoyBean_Tonnes'] + faostat['ARCD_Cattle_Beef_Tonnes'])/1e6, '--', color='black', label='Soy bean and cattle', linewidth=1)p3 = ax2.bar(t_y, datadict['EQAS_peatdrain'], align=align, color=ccat_fig1[2], edgecolor='white', linewidth=0.6, label='Peat drainage') # , hatch='.....')p4 = ax2.bar(t_y, datadict['EQAS_deco'], bottom=datadict['EQAS_peatdrain'], align=align, color=ccat_fig1[0], edgecolor='white', linewidth=0.6, label='Decomposition') # , hatch='')p5 = ax2.bar(t_y, (datadict['EQAS_fire'] + datadict['EQAS_peatfire']), bottom=(datadict['EQAS_peatdrain'] + datadict['EQAS_deco']), align=align, color=ccat_fig1[1], edgecolor='white', linewidth=0.6, label='Fire emissions') # , hatch='\\\\\\\\')# Add palmoil exportax21 = ax2.twinx() l3 = ax21.plot(t_y[2:], faostat['Indo_PalmOil_Tonnes']/1e6, '--', color='black', label='Palm oil', linewidth=1)ax1.set_xlim(1958, 2020)ax1.set_ylim(0, 0.9)ax1.set_yticks([0, 0.15, 0.3, 0.45, 0.6, 0.75, 0.9])ax1.grid()ax1.set_axisbelow(True)plt.setp(ax1.get_xticklabels(), visible=False)plt.setp(ax1.get_yticklabels(), fontsize=fontsize)ax11.set_ylim(0, 90)ax11.set_yticks([0,30,60,90])plt.setp(ax11.get_yticklabels(), fontsize=fontsize)handles = [p2, p1] + l1labels = [l.get_label() for l in handles]ax1.legend(handles, labels, loc='upper left', bbox_to_anchor=(0.0, 0.85), fontsize=fontsize_legend, framealpha=1)ax1.text(0.06, 0.9, 'a', horizontalalignment='right', verticalalignment='center', transform=ax1.transAxes, fontsize=fontsize_panellabel, weight='bold')ax1.set_ylabel('Emissions (Pg C year $^{-1}$)', size=fontsize)ax11.set_ylabel('Export commodities (Mt year $^{-1}$)', size=fontsize, rotation=270, va='bottom')ax1.yaxis.set_label_coords(-0.1,-0.1)ax11.yaxis.set_label_coords(1.075,-0.1)ax2.set_xlim(1958, 2020)ax2.set_xlabel('Year', fontsize=fontsize)ax2.set_ylim(0, 1.5)ax2.set_yticks([0, 0.25, 0.5, 0.75, 1.0, 1.25, 1.5])ax2.grid()ax2.set_axisbelow(True)plt.setp(ax2.get_xticklabels(), size=fontsize, visible=True)plt.setp(ax2.get_yticklabels(),fontsize=fontsize)ax21.set_ylim(0, 60)plt.setp(ax21.get_yticklabels(),fontsize=fontsize)handles = [p5, p4, p3]+l3labels = [l.get_label() for l in handles]ax2.legend(handles, labels, loc='upper left', bbox_to_anchor=(0.0, 0.85), fontsize=fontsize_legend, framealpha=1)ax2.text(0.06, 0.9, 'b', horizontalalignment='right', verticalalignment='center', transform=ax2.transAxes, fontsize=fontsize_panellabel, weight='bold')if filename:plt.savefig(filename + '.pdf', bbox_inches='tight')else:plt.show()
plot_Figure1(filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure1')# Determine annual trend for in Abstract text of manuscript:
LUnew_defos = datadict['Total'] - datadict['Other'] # Total deforestation emissions for EQAS and ARCD.
LUnew_defos_reg = slopest(t_y, LUnew_defos, method='linregres')
LUnew_defos_man = slopest(t_y, LUnew_defos, method='mannkendall')def plot_Figure2(fign, FF_China=None, filename=None):''' Plot paper Figure 2:param fign: figure number:param FF_China: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param filename: filename of saved figure (.pdf format). If filename == None, figure is only displayed and not saved.:return: Figure 2'''fontsize = 7fontsize_legend = 6.5fontsize_panellabel = 8if FF_China is None:FF_China = 'GCP'if FF_China == 'GCP': FF = np.array(FFgcp)elif FF_China == 'LIU': FF = np.array(FFadj)fig = plt.figure(fign, figsize=(183*mm, 89*mm))# Plot annual LUC emissionsax = fig.add_subplot(121)bottom = np.zeros(index1-index0+1)for keyn in metadict.keys():ax.bar(years, datadict[keyn], bottom=bottom, color=metadict[keyn][2], label=metadict[keyn][0], edgecolor='w', width=0.95) bottom += datadict[keyn]ax.text(0.03, 0.95, 'a', transform=ax.transAxes, fontsize=fontsize_panellabel, weight='bold')ax.set_xlim((year0, year1+1))ax.set_ylim((0, 2.5))ax.set_yticks((0,0.25,0.5,0.75,1.0,1.25,1.5,1.75,2.0,2.25,2.5))ax.set_xlabel('Year', fontsize=fontsize)ax.set_ylabel('Net LULCC emissions (Pg C year$^{-1}$)', fontsize=fontsize)ax.tick_params(axis='both', which='major', labelsize=fontsize)h,l = ax.get_legend_handles_labels()ax.legend(h[::-1], l[::-1], bbox_to_anchor=(0.01, 0.95), loc='upper left', fontsize=fontsize_legend, framealpha=1)ax.step(years+0.5, bottom, '-', color='lightgrey', linewidth=1.0)LULCC_this_study = bottom # == LUnew == datadict['Total']# Plot full carbon budgetax = fig.add_subplot(122)colors = tol_colors.tol_cset('vibrant')Cff = colors.greyCaccent = colors.tealax.step(years, FF, color=Cff, linewidth=1.5, label='Fossil fuel emissions') # 'grey'ax.step(years, dCO2, color=Caccent ,linewidth=1.5, label='Atmospheric CO$_2$ growth') # 'deepskyblue'ax.step(years, LUgcp, color=Cgcp, linewidth=1.5, label='LULCC GCP')ax.step(years, LUhan, color=Chan, linewidth=1.5, label='LULCC H&N')ax.step(years, LULCC_this_study, color=Cnew, linewidth=1.5, label='LULCC this study')# add ppm CO2 values as secondary axis.for i in np.arange(0.5, 4, 0.5):ax.text(year1+2, i * fPG, str(i), color=Caccent, va='center', fontsize=fontsize)ax.hlines(i * fPG, year1, year1+1, color=Caccent, linewidth=1.0)ax.text(year1+7, 2 * fPG, 'ppm CO$_2$ year$^{-1}$', rotation=270, color=Caccent, va='center', fontsize=fontsize)ax.text(0.03, 0.95, 'b', transform=ax.transAxes, fontsize=fontsize_panellabel, weight='bold')ax.set_xlim((year0, year1+1))ax.set_ylim((0, 11))ax.set_xlabel('Year', fontsize=fontsize)ax.set_ylabel('Pg C year$^{-1}$', fontsize=fontsize)ax.tick_params(axis='both', which='major', labelsize=fontsize)ax.legend(bbox_to_anchor=(0.01, 0.95), loc='upper left', fontsize=fontsize_legend, framealpha=1)plt.tight_layout()plt.subplots_adjust(left=0.075, right=0.94)if filename:plt.savefig(filename + '.pdf', bbox_inches='tight')else:plt.show()
plot_Figure2(2, filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure2')def calc_AF(lustr, FF_China=None, RESPscheme=None): #, trendcalc=None):''' Calculation of the Airborne fraction (without Monte-Carlo simulation).AF_a = annual raw, AF_ms = monthly smoothed, AF_msnv = monthly smoothed and filtered, AF_asnv = annual smoothed and filtered.:param lustr: 'gcp' (Global Carbon Project), 'han' (Houghton & Nassikas), or 'new' (This study):param FF_China: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param RESPscheme: choose respiration scheme, normal ('stable') or increasing role of fire ('incfire'):return: AF time series, AF error time series, and AF regression results.=== All results are returned for four data treatments ===:0 = annual raw ('AF_a') [Reported in paper Figure 3]1 = monthly smoothed ('AF_ms') [Not used in paper]2 = monthly smoothed and filtered ('AF_msnv') [Not used in paper]3 = annual smoothed and filtered ('AF_asnv') [Used for final trend estimates, reported in Table 1, Figure 3 and 4]'''if FF_China is None: FF_China = 'GCP'if RESPscheme is None: RESPscheme = 'stable'# Select LULCC and FF emissions time series:lu_tuple, ff_tuple = data_selector(lustr, FF_China, RESPscheme)lu, lu_m, lu_error, lu_error_m = lu_tupleFF, FF_m, FF_error, FF_error_m = ff_tuple# Calculate Airborne fractionAF_a = dCO2 / (FF + lu) # annual rawAF_ms = dCO2_s / (FF_m + lu_m) # monthly smoothedAF_msnv = dCO2_sv_m / (FF_m + lu_m) # monthly smoothed and filteredAF_asnv = dCO2_sv / (FF + lu) # annual smoothed and filteredAF_a_error = np.sqrt(dCO2_error**2 * (1/(FF + lu))**2 + (FF_error**2 + lu_error**2) * (-dCO2/(FF + lu)**2)**2) # Calculate error propagation.AF_ms_error = np.sqrt(dCO2_error_m**2 * (1/(FF_m + lu_m))**2 + (FF_error_m**2 + lu_error_m**2) * (-dCO2_m/(FF_m + lu_m)**2)**2)AF_msnv_error = np.sqrt(dCO2_error_m**2 * (1/(FF_m + lu_m))**2 + (FF_error_m**2 + lu_error_m**2) * (-dCO2_m/(FF_m + lu_m)**2)**2)AF_asnv_error = np.sqrt(dCO2_error**2 * (1/(FF + lu))**2 + (FF_error**2 + lu_error**2) * (-dCO2/(FF + lu)**2)**2)# Calculate trend in Airborne fraction time series.if trendcalc == 'abs': # Calculate absolute trend.AF_a_reg = slopest(t_y, AF_a, method=trendmethod)AF_ms_reg = slopest(t_m[edge:-edge], AF_ms[edge:-edge], method=trendmethod)AF_msnv_reg = slopest(t_m[edge:-edge], AF_msnv[edge:-edge], method=trendmethod)AF_asnv_reg = slopest(t_y, AF_asnv, method=trendmethod)elif trendcalc == 'rgr': # Calculate relative growth rate.AF_a_reg = slopest(t_y, AF_a / float(np.mean(AF_a)), method=trendmethod)AF_ms_reg = slopest(t_m[edge:-edge], AF_ms[edge:-edge] / np.mean(AF_ms[edge:-edge]), method=trendmethod)AF_msnv_reg = slopest(t_m[edge:-edge], AF_msnv[edge:-edge] / np.mean(AF_msnv[edge:-edge]), method=trendmethod)AF_asnv_reg = slopest(t_y, AF_asnv / float(np.mean(AF_asnv)), method=trendmethod)# Store regression results. Format: [slope, p-value, standard error]AF_a_reg = [AF_a_reg[0] * uconv, AF_a_reg[3] * 1, AF_a_reg[4] * uconv]AF_ms_reg = [AF_ms_reg[0] * uconv, AF_ms_reg[3] * 1, AF_ms_reg[4] * uconv]AF_msnv_reg = [AF_msnv_reg[0] * uconv, AF_msnv_reg[3] * 1, AF_msnv_reg[4] * uconv]AF_asnv_reg = [AF_asnv_reg[0] * uconv, AF_asnv_reg[3] * 1, AF_asnv_reg[4] * uconv]return (AF_a, AF_ms, AF_msnv, AF_asnv), (AF_a_error, AF_ms_error, AF_msnv_error, AF_asnv_error), (AF_a_reg, AF_ms_reg, AF_msnv_reg, AF_asnv_reg)
AFgcp, AFERRgcp, REGgcp = calc_AF('gcp') # AF: time series, AFERR: error time series, and REG: regression results.
AFhan, AFERRhan, REGhan = calc_AF('han')
AFnew, AFERRnew, REGnew = calc_AF('new')
AFnew_b, AFERRnew_b, REGnew_b = calc_AF('new', FF_China='LIU', RESPscheme='stable') # Different scenarios for sensitivity analysis.
AFnew_c, AFERRnew_c, REGnew_c = calc_AF('new', FF_China='GCP', RESPscheme='incfire')
AFnew_d, AFERRnew_d, REGnew_d = calc_AF('new', FF_China='LIU', RESPscheme='incfire')# # Save AF time series into Excel file
# sheet_AF = np.array([AFnew[0], AFERRnew[0], AFnew[3], AFERRnew[3], AFhan[0], AFERRhan[0], AFhan[3], AFERRhan[3], AFgcp[0], AFERRgcp[0], AFgcp[3], AFERRgcp[3]])
# pd.DataFrame(sheet_AF.T).to_excel(wdir+'Excel_AF_annual_2020.xlsx')
# sheet_AFmonthly = np.array([AFnew[1], AFERRnew[1], AFnew[2], AFERRnew[2], AFhan[1], AFERRhan[1], AFhan[2], AFERRhan[2], AFgcp[1], AFERRgcp[1], AFgcp[2], AFERRgcp[2]])
# pd.DataFrame(sheet_AFmonthly.T).to_excel(wdir+'Excel_AF_monthly_2020.xlsx')def calc_AF_MonteCarlo(MC, lustr, FF_China=None, RESPscheme=None, bootstrap=False, detrend=False):''' Calculation of the Airborne fraction using Monte Carlo simulation.AF_a = annual raw, AF_ms = monthly smoothed, AF_msnv = monthly smoothed and filtered, AF_asnv = annual smoothed and filtered.:param MC: number of Monte Carlo iterations.:param lustr: 'gcp' (Global Carbon Project), 'han' (Houghton & Nassikas), or 'new' (This study):param FF_China: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param RESPscheme: choose respiration scheme, normal ('stable', default) or increasing role of fire ('incfire'):param bootstrap: Apply bootstrapping for calculation of trend uncertainty interval (default=False).:param detrend: Remove trend before calculation, to isolate the error margin around 0 (default=False).:return:- MC_slope_median: median of found slopes over MonteCarlo iterations.- MC_slope_std: standard deviation of found slopes over MonteCarlo iterations.- MC_std_mean: mean of found standard deviations over MonteCarlo iterations.- MC_p_mean: mean of found p-values over MonteCarlo iterations.- MC_slope: Found slopes for all MonteCarlo iterations.- MC_std: Found standard deviations for all MonteCarlo iterations.- MC_p: Found p-values for all MonteCarlo iterations.- MC_AF: Total AF time series for all MonteCarlo iterations.- MC_interc: Found intercept for all MonteCarlo iterations.=== All results are returned for four data treatments ===:0 = annual raw ('AF_a') [Reported in paper Figure 3]1 = monthly smoothed ('AF_ms') [Not used in paper]2 = monthly smoothed and filtered ('AF_msnv') [Not used in paper]3 = annual smoothed and filtered ('AF_asnv') [Used for final trend estimates, reported in Table 1, Figure 3 and 4]'''if FF_China is None: FF_China = 'GCP'if RESPscheme is None: RESPscheme = 'stable'if bootstrap is True:fBts = 0.8 # Bootstrap window of at least 80% of time series.min_length_y = int(np.ceil(len(years) * fBts)) # at least fBts.min_length_m = int(np.ceil((len(t_m) - mavg) * fBts)) # at least fBts.start = np.zeros(MC).astype(int)length = np.zeros(MC).astype(int)end = np.zeros(MC).astype(int)# Select used land use and fossil fuel time serieslu_tuple, ff_tuple = data_selector(lustr, FF_China, RESPscheme)lu, lu_m, lu_error, lu_error_m = lu_tupleFF, FF_m, FF_error, FF_error_m = ff_tupleAF_pre = calc_AF(lustr, FF_China=FF_China, RESPscheme=RESPscheme)[0]AF_a_trend = np.poly1d(np.polyfit(t_y, AF_pre[0], 1))(t_y) # Determine Airborne fraction trend component.AF_ms_trend = np.poly1d(np.polyfit(t_m, AF_pre[1], 1))(t_m)AF_msnv_trend = np.poly1d(np.polyfit(t_m, AF_pre[2], 1))(t_m)AF_asnv_trend = np.poly1d(np.polyfit(t_y, AF_pre[3], 1))(t_y)print('Monte Carlo starting ... ')time0 = timer.time()AF_a = np.zeros((MC, len(years)))AF_ms = np.zeros((MC, len(t_m)))AF_msnv = np.zeros((MC, len(t_m)))AF_asnv = np.zeros((MC, len(years)))MC_a = np.zeros((4, MC)) # 4 dimensions are: [slope, intercept, p-value, stderr]MC_ms = np.zeros((4, MC))MC_msnv = np.zeros((4, MC))MC_asnv = np.zeros((4, MC))for mc in range(MC):FF_mc = FF + FF_error * np.random.randn(len(years)) # Apply random normally-distributed error.lu_mc = lu + lu_error * np.random.randn(len(years))dCO2_mc = dCO2 + dCO2_error * np.random.randn(len(years))FF_m_mc = FF_m + FF_error_m * np.random.randn(len(t_m))lu_m_mc = lu_m + lu_error_m * np.random.randn(len(t_m))dCO2_m_mc = dCO2_m + dCO2_error_m * np.random.randn(len(t_m))dCO2_s_mc = dCO2_s + dCO2_error_m * np.random.randn(len(t_m))dCO2_sv_mc = dCO2_sv + dCO2_error * np.random.randn(len(years))dCO2_sv_m_mc = dCO2_sv_m + dCO2_error_m * np.random.randn(len(t_m))# Calculate Airborne fractionAF_a[mc] = dCO2_mc / (FF_mc + lu_mc) # annual rawAF_ms[mc] = dCO2_s_mc / (FF_m_mc + lu_m_mc) # monthly smoothedAF_msnv[mc] = dCO2_sv_m_mc / (FF_m_mc + lu_m_mc) # monthly smoothed and filteredAF_asnv[mc] = dCO2_sv_mc / (FF_mc + lu_mc) # annual smoothed and filteredif detrend is True:AF_a[mc] = AF_a[mc] - AF_a_trend # Remove time series trend.AF_ms[mc] = AF_ms[mc] - AF_ms_trendAF_msnv[mc] = AF_msnv[mc] - AF_msnv_trendAF_asnv[mc] = AF_asnv[mc] - AF_asnv_trendif bootstrap is True:length_y = np.random.randint(min_length_y, len(years)+1) # bootstrap window lengthstart_y = np.random.randint(len(years) - length_y+1) # bootstrap window start pointend_y = start_y + length_y # bootstrap window end pointlength_m = np.random.randint(min_length_m, len(t_m) - mavg +1) # Same, but monthly instead of annualstart_m = np.random.randint(edge, len(t_m) - edge - length_m +1)end_m = start_m + length_mmask_y = np.zeros(len(years)).astype(bool)mask_m = np.zeros(len(t_m)).astype(bool)mask_y[start_y:end_y] = True # Bootstrap window mask, annual.mask_m[start_m:end_m] = True # Bootstrap window mask, monthly.AF_a[mc][~mask_y] = np.nan # Apply bootstrap mask to AF time series.AF_ms[mc][~mask_m] = np.nanAF_msnv[mc][~mask_m] = np.nanAF_asnv[mc][~mask_y] = np.nanlength[mc] = length_m # store iterated bootstrap windows.start[mc] = start_mend[mc] = end_melif bootstrap is False: # Make empty mask to skip bootstrapping.mask_y = np.zeros(len(years)).astype(bool)mask_m = np.zeros(len(t_m)).astype(bool)mask_y[:] = Truemask_m[edge:-edge] = True# Calculate trend in Airborne fraction time series.if trendcalc == 'abs': # Calculate absolute trend.AF_a_reg = slopest(t_y[mask_y], AF_a[mc][mask_y], method=trendmethod)AF_ms_reg = slopest(t_m[mask_m], AF_ms[mc][mask_m], method=trendmethod)AF_msnv_reg = slopest(t_m[mask_m], AF_msnv[mc][mask_m], method=trendmethod)AF_asnv_reg = slopest(t_y[mask_y], AF_asnv[mc][mask_y], method=trendmethod)elif trendcalc == 'rgr': # Calculate relative growth rate.AF_a_reg = slopest(t_y[mask_y], AF_a[mc][mask_y] / float(np.mean(AF_a[mc][mask_y])), method=trendmethod)AF_ms_reg = slopest(t_m[mask_m], AF_ms[mc][mask_m] / np.mean(AF_ms[mc][mask_m]), method=trendmethod)AF_msnv_reg = slopest(t_m[mask_m], AF_msnv[mc][mask_m] / np.mean(AF_msnv[mc][mask_m]), method=trendmethod)AF_asnv_reg = slopest(t_y[mask_y], AF_asnv[mc][mask_y] / float(np.mean(AF_asnv[mc][mask_y])), method=trendmethod)# Store regression results. Format: [slope, intercept, p-value, standard error]MC_a[:,mc] = [AF_a_reg[0]*uconv, AF_a_reg[1]*uconv, AF_a_reg[3]*1, AF_a_reg[4]*uconv]MC_ms[:,mc] = [AF_ms_reg[0]*uconv, AF_ms_reg[1]*uconv, AF_ms_reg[3]*1, AF_ms_reg[4]*uconv]MC_msnv[:,mc] = [AF_msnv_reg[0]*uconv, AF_msnv_reg[1]*uconv, AF_msnv_reg[3]*1, AF_msnv_reg[4]*uconv]MC_asnv[:,mc] = [AF_asnv_reg[0]*uconv, AF_asnv_reg[1]*uconv, AF_asnv_reg[3]*1, AF_asnv_reg[4]*uconv]# Calculate summary statistics over Monte Carlo iterations.MC_AF = (AF_a, AF_ms, AF_msnv, AF_asnv)MC_slope = np.array([MC_a[0], MC_ms[0], MC_msnv[0], MC_asnv[0]])MC_interc = np.array([MC_a[1], MC_ms[1], MC_msnv[1], MC_asnv[1]])MC_std = np.array([MC_a[3], MC_ms[3], MC_msnv[3], MC_asnv[3]])MC_p = np.array([MC_a[2], MC_ms[2], MC_msnv[2], MC_asnv[2]])MC_slope_median = np.mean(MC_slope, axis=1) # median of slopeMC_slope_std = np.std(MC_slope, axis=1) # std of slopeMC_std_mean = np.mean(MC_std, axis=1) # mean of stderrMC_p_mean = np.mean(MC_p, axis=1) # mean of p-valuetime1 = timer.time() - time0print('Done, duration = ' + str(time1))return MC_slope_median, MC_slope_std, MC_std_mean, MC_p_mean, MC_slope, MC_std, MC_p, MC_interc, MC_AF[0], MC_AF[1], MC_AF[2], MC_AF[3]def calc_AF_MonteCarlo_filemanager(filename):''' Saves or loads Monte-Carlo results for all different data scenarios.If zip file with filename already exists, previously saved results are loaded.If no zip file exists yet, the Monte-Carlo simulations are calculated for all different scenarios and saved into a zip file.:param filename: filename without path and without extension.:return: MCdict, dictionary collection with all run results.'''if (filename is not None) and os.path.isfile(filename + '.zip'):# Load pre-saved restuls.MCdict = OrderedDict()with ZipFile(filename + '.zip', mode='r') as archive:for keyn in ['gcp', 'han', 'new', 'gcp_s', 'han_s', 'new_s', 'new_b', 'new_c', 'new_d', 'new_e', 'new_f', 'gcp_dt', 'han_dt', 'new_dt']:ds_npz = np.load(io.BytesIO(archive.read(keyn+'.npz')))MCdict[keyn] = [ds_npz[filen] for filen in ['MC_slope_median', 'MC_slope_std', 'MC_std_mean', 'MC_p_mean', 'MC_slope', 'MC_std', 'MC_p', 'MC_interc', 'AF_a', 'AF_ms', 'AF_msnv', 'AF_asnv']]else:raise Warning('No pre-saved .zip file found with the name: %s, recalculating Monte-Carlo, Please note: this can take several minutes!' % filename)# Calculate and save results.MCdict = OrderedDict([('gcp', calc_AF_MonteCarlo(MC, 'gcp')),('han', calc_AF_MonteCarlo(MC, 'han')),('new', calc_AF_MonteCarlo(MC, 'new')),('gcp_s', calc_AF_MonteCarlo(MC, 'gcp', bootstrap=True)),('han_s', calc_AF_MonteCarlo(MC, 'han', bootstrap=True)),('new_s', calc_AF_MonteCarlo(MC, 'new', bootstrap=True)),('new_b', calc_AF_MonteCarlo(MC, 'new', FF_China='LIU', RESPscheme='stable', bootstrap=True)),('new_c', calc_AF_MonteCarlo(MC, 'new', FF_China='GCP', RESPscheme='incfire', bootstrap=True)),('new_d', calc_AF_MonteCarlo(MC, 'new', FF_China='LIU', RESPscheme='incfire', bootstrap=True)),('new_e', calc_AF_MonteCarlo(MC, 'new', FF_China='BP', RESPscheme='stable', bootstrap=True)),('new_f', calc_AF_MonteCarlo(MC, 'new', FF_China='BP', RESPscheme='incfire', bootstrap=True)),('gcp_dt', calc_AF_MonteCarlo(MC, 'gcp', detrend=True)),('han_dt', calc_AF_MonteCarlo(MC, 'han', detrend=True)),('new_dt', calc_AF_MonteCarlo(MC, 'new', detrend=True)),])# Convert dtype from Float64 to Float32 for reduced file size.for keyn in MCdict.keys():MCdict[keyn] = [dicti.astype(np.float32) for dicti in MCdict[keyn]]# Save results into .npz files and then pack them all into one .zip file.for keyn in MCdict.keys():ds = MCdict[keyn]np.savez(filename + '_%s' % keyn, MC_slope_median=ds[0], MC_slope_std=ds[1], MC_std_mean=ds[2], MC_p_mean=ds[3], MC_slope=ds[4], MC_std=ds[5], MC_p=ds[6], MC_interc=ds[7], AF_a=ds[8], AF_ms=ds[9], AF_msnv=ds[10], AF_asnv=ds[11])zipObj = ZipFile(filename + '.zip', 'w')for keyn in MCdict.keys():zipObj.write(filename + '_%s.npz' % keyn, keyn+'.npz') # Pack individual results into single .zip file.os.remove(filename + '_%s.npz' % keyn) # Remove individual files.zipObj.close()return MCdictMC = 10000 # number of Monte Carlo iterations.
MCdict = calc_AF_MonteCarlo_filemanager(wdir + 'Marle_et_al_Nature_AirborneFraction_MC%s_MK_ts_TREND%s' % (MC, trendcalc))def plot_Figure3(fign, lustr, FFchinaplot='LIU', filename=None):''' Plot paper Figure 3:param fign: figure number:param lustr: 'gcp' (Global Carbon Project), 'han' (Houghton & Nassikas), or 'new' (This study):param FFchinaplot: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param filename: filename of saved figure (.pdf format). If no filename, figure is only displayed and not saved.:return: Figure 3'''fontsize = 8fontsize_legend = 6.5fontsize_boxtext = 7fontsize_panellabel = 8fig = plt.figure(fign, (183*mm,70*mm))ax = fig.add_subplot(121)if lustr == 'gcp':AFdat = AFgcpMCdat = MCdict['gcp']MCdats = MCdict['gcp_s']AFERRdat = AFERRgcpAFcol = CgcpAFstr = 'GCP'pltlabels = ['a','b']elif lustr == 'han':AFdat = AFhanMCdat = MCdict['han']MCdats = MCdict['han_s']AFERRdat = AFERRhanAFcol = ChanAFstr = 'H&N'pltlabels = ['c', 'd']elif lustr == 'new':AFdat = AFnewMCdat = MCdict['new']MCdats = MCdict['new_s']AFERRdat = AFERRnewAFcol = CnewAFstr = 'This study'pltlabels = ['e', 'f']t_plot = t_yt_len = len(years)pl1, = ax.plot(t_plot, amean(AFdat[0]), c='grey') # Plot AF time seriespl2, = ax.plot(t_plot, amean(AFdat[iset]), c=AFcol)if lustr in ['gcp', 'new']: colorfillalpha = 0.2elif lustr == 'han': colorfillalpha = 0.3 # The yellow colors requires a slightly higher alpha for the same visual effect.ax.fill_between(t_plot, amean(AFdat[0] - AFERRdat[0]), amean(AFdat[0] + AFERRdat[0]), facecolor='grey', alpha=0.2) # Plot AF uncertainty rangeax.fill_between(t_plot, amean(AFdat[iset] - AFERRdat[iset]), amean(AFdat[iset] + AFERRdat[iset]), facecolor=AFcol, alpha=colorfillalpha)ax.plot(t_plot, np.poly1d(np.polyfit(t_plot, amean(AFdat[iset]),1))(t_plot), c=AFcol) # Plot AF trend lineMC_mask = np.where(~np.isnan(MCdats[8+iset]), 1, MCdats[8+iset]) # Remove bootstrap no-data edges.MC_line = (MCdats[4][iset] * (t_plot * MC_mask).T + MCdats[7][iset]).T / uconvMC_linemean = np.nanmedian(MC_line, axis=0)MC_linestd = np.nanstd(MC_line, axis=0)MC_linemean_fit = np.poly1d(np.polyfit(t_plot[2:-2], MC_linemean[2:-2], 3))(t_plot)MC_linestd_fit = np.poly1d(np.polyfit(t_plot[2:-2], MC_linestd[2:-2], 3))(t_plot)if lustr in ['gcp', 'han']: dashlinealpha = 0.7elif lustr == 'new': dashlinealpha = 0.9pl3, = ax.plot(t_plot, MC_linemean_fit - MC_linestd_fit, color=AFcol, linestyle='--', alpha=dashlinealpha) # Plot AF trend line uncertaintyax.plot(t_plot, MC_linemean_fit + MC_linestd_fit, color=AFcol, linestyle='--', alpha=dashlinealpha)p_dat = MCdat[3][iset]if lustr in ['gcp', 'han']:Pdat = (np.sum([MCdat[4][iset] > 0.0])/float(MC)) # Probability that trend is positivePdat2 = (np.sum([MCdat[6][iset] < 0.05])/float(MC)) # Probability that trend is significantPdats = (np.sum([MCdats[4][iset] > 0.0])/float(MC)) # Probability that trend is positivePdats2 = (np.sum([MCdats[6][iset] < 0.05])/float(MC)) # Probability that trend is significantelif lustr == 'new':Pdat = (np.sum([MCdat[4][iset] < 0.0])/float(MC)) # Probability that trend is negativePdat2 = (np.sum([MCdat[6][iset] < 0.05])/float(MC)) # Probability that trend is significantPdats = (np.sum([MCdats[4][iset] < 0.0])/float(MC)) # Probability that trend is negativePdats2 = (np.sum([MCdats[6][iset] < 0.05])/float(MC)) # Probability that trend is significantdata_list = [MCdict['gcp'][4][iset], MCdict['han'][4][iset], MCdict['new'][4][iset], MCdict['gcp_dt'][4][iset], MCdict['han_dt'][4][iset], MCdict['new_dt'][4][iset]]pval = np.zeros((6,6))# Example: pval = scipy.stats.t.sf(np.abs(3.0914), 8)*2 (Source: https://www.youtube.com/watch?v=myL_qzuLHTQ)for i, Di in enumerate(data_list):for j, Dj in enumerate(data_list):diff_slope = np.mean(Di) - np.mean(Dj)diff_std = np.sqrt(np.std(Di) ** 2 + np.std(Dj) ** 2)pval[i,j] = scipy.stats.t.sf(np.abs(diff_slope / diff_std), t_len - 4) * 2if trendcalc == 'rgr':spacer = 0.02legtext1 = '%s, raw. Mean AF = %.2f' % (AFstr, np.mean(AFdat[0]))legtext2 = '%s: %.2f $\pm$ %.2f %% yr$^{-1}$' % ('Filter', MCdat[0][iset], MCdat[1][iset])legtext3 = '%s: %.2f $\pm$ %.2f %% yr$^{-1}$' % ('Filter+Bootstrap', MCdats[0][iset], MCdats[1][iset])elif trendcalc == 'abs':spacer = 0.001legtext1 = '%s, raw, Mean AF = %.2f' % (AFstr, np.mean(AFdat[0]))legtext2 = '%s: %.3f $\pm$ %.3f decade$^{-1}$' % ('Filter', MCdat[0][iset], MCdat[1][iset])legtext3 = '%s: %.3f $\pm$ %.3f decade$^{-1}$' % ('Filter+Bootstrap', MCdats[0][iset], MCdats[1][iset])ax.legend([pl1,pl2,pl3], [legtext1, legtext2, legtext3], fontsize=fontsize_legend, loc='upper right', bbox_to_anchor=(1.01,1.02), labelspacing=0.05, borderpad=0.25, handlelength=1.5, framealpha=1)ax.text(0.04, 0.93, pltlabels[0], transform=ax.transAxes, fontsize=fontsize_panellabel, weight='bold')ax.set_xlabel('Year', fontsize=fontsize)ax.set_ylabel('Airborne fraction (unitless)', fontsize=fontsize)ax.set_xlim([year0-1, year1+1])ax.set_ylim([0.1,0.9])ax.grid(True)ax.set_axisbelow(True)ax.tick_params(axis='both', which='major', labelsize=fontsize)ax = fig.add_subplot(122)width_bp = 0.8if lustr in ['gcp', 'han']:bp1 = ax.boxplot(MCdat[4][0], positions=[4], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp2 = ax.boxplot(MCdat[4][iset], positions=[3], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp3 = ax.boxplot(MCdats[4][0], positions=[2], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp4 = ax.boxplot(MCdats[4][iset], positions=[1], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])for b, bplot in enumerate((bp1, bp2, bp3, bp4)):bplot['boxes'][0].set(facecolor='white')if b in [0,2]:bplot['boxes'][0].set(edgecolor=rgba2rgb(np.array(pltc.to_rgba(AFcol, 0.3))*255)/255.)ylim = [0.0, 5]bploc_wisk = np.reshape([item.get_xdata()[1] for bp in [bp1, bp2, bp3, bp4] for item in bp['whiskers']], (4, 2)) # [:,0] is left, [:,1] is right.bploc_box = np.reshape([item.get_xdata()[0] for bp in [bp1, bp2, bp3, bp4] for item in bp['whiskers']], (4, 2)) # [:,0] is left, [:,1] is right.if lustr == 'gcp':ax.text(bploc_wisk[0, 0] - spacer, 4, 'Raw', color='grey', va='center', ha='right', bbox=dict(facecolor='white', pad=0, edgecolor='none'), zorder=100, fontsize=fontsize_boxtext)ax.text(bploc_wisk[1, 0] - spacer, 3, 'Filter', color='grey', va='center', ha='right', bbox=dict(facecolor='white', pad=2, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[2, 0] - spacer, 2, 'Bootstrap', color='grey', va='center', ha='right', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[3, 0] - spacer, 1, 'Filter+Bootstrap', color='grey', va='center', ha='right', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)elif lustr == 'han':ax.text(bploc_wisk[0, 1] + spacer, 4, 'Raw', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), zorder=100, fontsize=fontsize_boxtext)ax.text(bploc_wisk[1, 1] + spacer, 3, 'Filter', color='grey', va='center', bbox=dict(facecolor='white', pad=2, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[2, 1] + spacer, 2, 'Bootstrap', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[3, 1] + spacer, 1, 'Filter+Bootstrap', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)elif lustr == 'new':bp1 = ax.boxplot(MCdat[4][0], positions=[8], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5, 95])bp2 = ax.boxplot(MCdat[4][iset], positions=[7], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5, 95])bp3 = ax.boxplot(MCdats[4][0], positions=[6], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5, 95])bp4 = ax.boxplot(MCdats[4][iset], positions=[5], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5, 95])if FFchinaplot == 'LIU':bp5 = ax.boxplot(MCdict['new_b'][4][iset], positions=[3], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp6 = ax.boxplot(MCdict['new_c'][4][iset], positions=[2], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp7 = ax.boxplot(MCdict['new_d'][4][iset], positions=[1], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])elif FFchinaplot == 'BP':bp5 = ax.boxplot(MCdict['new_e'][4][iset], positions=[3], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp6 = ax.boxplot(MCdict['new_c'][4][iset], positions=[2], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])bp7 = ax.boxplot(MCdict['new_f'][4][iset], positions=[1], vert=False, sym='', widths=width_bp, patch_artist=True, boxprops=dict(color=AFcol), whis=[5,95])for b, bplot in enumerate((bp1, bp2, bp3, bp4, bp5, bp6, bp7)):bplot['boxes'][0].set(facecolor='white')if b in [0,2]:bplot['boxes'][0].set(edgecolor=rgba2rgb(np.array(pltc.to_rgba(AFcol, 0.3))*255)/255.)ylim = [-.05, 9]bploc_wisk = np.reshape([item.get_xdata()[1] for bp in [bp1, bp2, bp3, bp4, bp5, bp6, bp7] for item in bp['whiskers']], (7, 2)) # [:,0] is left, [:,1] is right.bploc_box = np.reshape([item.get_xdata()[0] for bp in [bp1, bp2, bp3, bp4, bp5, bp6, bp7] for item in bp['whiskers']], (7, 2)) # [:,0] is left, [:,1] is right.ax.text(bploc_wisk[0, 1] + spacer, 8, 'Raw', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), zorder=100, fontsize=fontsize_boxtext)ax.text(bploc_wisk[1, 1] + spacer, 7, 'Filter', color='grey', va='center', bbox=dict(facecolor='white', pad=2, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[2, 1] + spacer, 6, 'Bootstrap', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[3, 1] + spacer, 5, 'Filter+Bootstrap', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[4, 1] + spacer, 3, 'Lower FF emissions China', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), zorder=100, fontsize=fontsize_boxtext)ax.text(bploc_wisk[5, 1] + spacer, 2, 'Increasing role of fire', color='grey', va='center', bbox=dict(facecolor='white', pad=2, edgecolor='none'), fontsize=fontsize_boxtext)ax.text(bploc_wisk[6, 1] + spacer, 1, 'Combined adjustments', color='grey', va='center', bbox=dict(facecolor='white', pad=0, edgecolor='none'), fontsize=fontsize_boxtext)ax.axvline(x=0, c='black', alpha=0.6, zorder=0)ax.set_xlim([-0.035,0.035])ax.get_yaxis().set_ticks([])ax.set_ylim(ylim)ax.grid(True)ax.set_axisbelow(True)ax.tick_params(axis='both', which='major', labelsize=fontsize)if lustr == 'gcp':textstr = 'GCP vs. H&N: p = %.2f\nGCP vs. This study: p = %.2f\nGCP vs. no trend: p = %.2f' % (pval[0,1], pval[0,2], pval[0,3])elif lustr == 'han':textstr = 'H&N vs. GCP: p = %.2f\nH&N vs. This study: p = %.2f\nH&N vs. no trend: p = %.2f' % (pval[0,1], pval[1,2], pval[1,4])elif lustr == 'new':textstr = 'This study vs. GCP: p = %.2f\nThis study vs. H&N: p = %.2f\nThis study vs. no trend: p = %.2f' % (pval[0,2], pval[1,2], pval[2,5])ax.text(0.015, 0.93, pltlabels[1], transform=ax.transAxes, fontsize=fontsize_panellabel, weight='bold')if trendcalc == 'rgr':ax.set_xlabel('Relative growth rate, RGR (% yr$^{-1}$)', fontsize=fontsize)elif trendcalc == 'abs':ax.set_xlabel('Trend (decade$^{-1}$)', fontsize=fontsize)plt.tight_layout()if filename:plt.savefig(filename + '.pdf', bbox_inches='tight')else:plt.show()
plot_Figure3(3, lustr='gcp', filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure3_MC%s_ab' % MC)
plot_Figure3(4, lustr='han', filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure3_MC%s_cd' % MC)
plot_Figure3(5, lustr='new', filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure3_MC%s_ef' % MC)def plot_Table1():''' Calculate Table 1 from paper with trend significance values.:return: Table 1, dataframe containing significance values.'''datalist = [MCdict['gcp'], MCdict['han'], MCdict['new'], MCdict['gcp_s'], MCdict['han_s'], MCdict['new_s']]Ppos = np.zeros(len(datalist))Pneg = np.zeros(len(datalist))P2 = np.zeros(len(datalist))Ppos2 = np.zeros(len(datalist))Pneg2 = np.zeros(len(datalist))for i, MCdat in enumerate(datalist):Ppos[i] = np.sum([MCdat[4][iset] > 0.0]) / float(MC) # Probability P of a positive trendPneg[i] = np.sum([MCdat[4][iset] < 0.0]) / float(MC) # Probability P of a negative trendP2[i] = np.sum([MCdat[6][iset] < 0.05])/float(MC) # Probability P of a significant trend (both positive and negative)Ppos2[i] = np.sum([(MCdat[4][iset] > 0.0) & (MCdat[6][iset] < 0.05)]) / float(MC) # Probability P of a significant positive trendPneg2[i] = np.sum([(MCdat[4][iset] < 0.0) & (MCdat[6][iset] < 0.05)]) / float(MC) # Probability P of a significant negative trenddframe = np.array([[Ppos[3],Pneg[3],Ppos2[3],Pneg2[3],Ppos2[3]/(Ppos2[3]+Pneg2[3]),Pneg2[3]/(Ppos2[3]+Pneg2[3])], # Store results in dataframe.[Ppos[4],Pneg[4],Ppos2[4],Pneg2[4],Ppos2[4]/(Ppos2[4]+Pneg2[4]),Pneg2[4]/(Ppos2[4]+Pneg2[4])],[Ppos[5],Pneg[5],Ppos2[5],Pneg2[5],Ppos2[5]/(Ppos2[5]+Pneg2[5]),Pneg2[5]/(Ppos2[5]+Pneg2[5])]])dframe = pd.DataFrame(dframe, index=['GCP','H&N','This study'], columns=['Positive','Negative','Positive p<0.05','Negative p<0.05','p<0.05 fraction positive','p<0.05 fraction negative'])return dframe
ds_table1 = plot_Table1()
print(ds_table1.to_string())def calc_AF_MonteCarlo_ARR(MC, size, filename=None, FF_China=None, RESPscheme=None, bootstrap=True):''' Calculation of the Airborne fraction using a Monte Carlo simulation, based on the linear approximations of the LU time series.For a (size+1 x size+1) array, the AF is calculated for a range of intercepts and slopes.Three additional intercepts and slopes are added; those are the linear fits of the gcp, han, and new LU time series.Those three end up in the extended diagonal of the results arrays. They can be extracted using indices [size+1,size+1], [size+2,size+2], and [size+3,size+3]AF_a = annual raw, AF_ms = monthly smoothed, AF_msnv = monthly smoothed and filtered, AF_asnv = annual smoothed and filtered.:param MC: number of Monte Carlo iterations.:param size: size of data array.:param filename: filename of saved result (saved in .npz format) [str]:param FF_China: choose which FF scheme to use, China GCP ('GCP'), China Liu et al. (2015) ('LIU'), or China BP (2021):param RESPscheme: choose respiration scheme, normal ('stable', default) or increasing role of fire ('incfire'):param bootstrap: Apply bootstrapping for calculation of trend uncertainty interval (default=True).:return:- MC_slope_median: median of found slopes over MonteCarlo iterations.- MC_slope_std: standard deviation of found slopes over MonteCarlo iterations.- MC_std_mean: mean of found standard deviations over MonteCarlo iterations.- MC_p_mean: mean of found p-values over MonteCarlo iterations.=== All results are returned for four data treatments ===:0 = annual raw ('AF_a') [Reported in paper Figure 3]1 = monthly smoothed ('AF_ms') [Not used in paper]2 = monthly smoothed and filtered ('AF_msnv') [Not used in paper]3 = annual smoothed and filtered ('AF_asnv') [Used for final trend estimates, reported in Table 1, Figure 3 and 4]'''if FF_China is None: FF_China = 'GCP'if RESPscheme is None: RESPscheme = 'stable'if bootstrap is True:fBts = 0.8 # Bootstrap window of at least 80% of time series.min_length_y = int(np.ceil(len(years) * fBts)) # at least fBts.min_length_m = int(np.ceil((len(t_m) - mavg) * fBts)) # at least fBts.start = np.zeros(MC).astype(int)length = np.zeros(MC).astype(int)end = np.zeros(MC).astype(int)if (filename is not None) and os.path.isfile(filename + '.npz'):ds = np.load(filename + '.npz')MC_slope_median = ds['MC_slope_median'][:]MC_slope_std = ds['MC_slope_std'][:]MC_std_mean = ds['MC_std_mean'][:]MC_p_mean = ds['MC_p_mean'][:]else:raise Warning('No pre-saved .npz file found with the name: %s, recalculating Monte-Carlo, Please note: this can take up to 30 hours!' % filename)# Select used land use and fossil fuel time series_, ff_tuple = data_selector('gcp', FF_China=FF_China, RESPscheme=RESPscheme) # 'gcp' is a placeholder here, not used. Only FF is loaded.FF, FF_m, FF_error, FF_error_m = ff_tupleLUgcp_slope, _ = np.polyfit(t_y, LUgcp, 1) # determine slope and b for lu datasetsLUhan_slope, _ = np.polyfit(t_y, LUhan, 1)LUnew_slope, _ = np.polyfit(t_y, LUnew, 1)LUgcp_mean = np.mean(LUgcp) # determine mean for lu datasetsLUhan_mean = np.mean(LUhan)LUnew_mean = np.mean(LUnew)LUgcp_std = np.std(LUgcp)LUhan_std = np.std(LUhan)LUnew_std = np.std(LUnew)a = np.linspace(-0.005, 0.025, size+1)a = np.hstack((a, LUgcp_slope, LUhan_slope, LUnew_slope)) # add 3 linear lines with slope of lu datasetsm = np.linspace(0.8, 1.5+(1/100.), size+1)m = np.hstack((m, LUgcp_mean, LUhan_mean, LUnew_mean))fsize = size+1+3 # plus 1 for symmetry, plus 3 for 3 LU datasets.mesha = np.zeros((fsize,fsize))meshm = np.zeros((fsize,fsize))MC_slope_median = np.zeros((4,fsize,fsize))MC_slope_std = np.zeros((4,fsize,fsize))MC_std_mean = np.zeros((4,fsize,fsize))MC_p_mean = np.zeros((4,fsize,fsize))for i in [0,51,52,53]: #range(fsize):for j in [0,51,52,53]: #range(fsize):print('i = '+str(i)+' / j = '+str(j) + ' ... ')time0 = timer.time()mesha[i,j] = a[i]meshm[i,j] = m[j]lu = a[i] * (np.arange(len(years)) - len(years)/2.0) + m[j]lu_m = tomonthly(lu) # create monthly series by interpolation of yearly serieslu_error = lu * 0.5 # LU errors are set at 50% for all three datasets.lu_error_m = lu_m * 0.5MC_a = np.zeros((4, MC)) # 4 dimensions are: [slope, intercept, p-value, stadard error]MC_ms = np.zeros((4, MC))MC_msnv = np.zeros((4, MC))MC_asnv = np.zeros((4, MC))for mc in range(MC):FF_mc = FF + FF_error * np.random.randn(len(years)) # Apply random normally-distributed error.lu_mc = lu + lu_error * np.random.randn(len(years))dCO2_mc = dCO2 + dCO2_error * np.random.randn(len(years))FF_m_mc = FF_m + FF_error_m * np.random.randn(len(t_m))lu_m_mc = lu_m + lu_error_m * np.random.randn(len(t_m))dCO2_m_mc = dCO2_m + dCO2_error_m * np.random.randn(len(t_m))dCO2_s_mc = dCO2_s + dCO2_error_m * np.random.randn(len(t_m))dCO2_sv_mc = dCO2_sv + dCO2_error * np.random.randn(len(years))dCO2_sv_m_mc = dCO2_sv_m + dCO2_error_m * np.random.randn(len(t_m))# Calculate Airborne fractionAF_a = dCO2_mc / (FF_mc + lu_mc) # annual rawAF_ms = dCO2_s_mc / (FF_m_mc + lu_m_mc) # monthly smoothedAF_msnv = dCO2_sv_m_mc / (FF_m_mc + lu_m_mc) # monthly smoothed and filteredAF_asnv = dCO2_sv_mc / (FF_mc + lu_mc) # annual smoothed and filteredif bootstrap is True:length_y = np.random.randint(min_length_y, len(years)+1) # bootstrap window lengthstart_y = np.random.randint(len(years) - length_y+1) # bootstrap window start pointend_y = start_y + length_y # bootstrap window end pointlength_m = np.random.randint(min_length_m, len(t_m) - mavg +1) # Same, but monthly instead of annualstart_m = np.random.randint(edge, len(t_m) - edge - length_m +1)end_m = start_m + length_mmask_y = np.zeros(len(years)).astype(bool)mask_m = np.zeros(len(t_m)).astype(bool)mask_y[start_y:end_y] = True # Bootstrap window mask, annual.mask_m[start_m:end_m] = True # Bootstrap window mask, monthly.AF_a[~mask_y] = np.nan # Apply bootstrap mask to AF time series.AF_ms[~mask_m] = np.nanAF_msnv[~mask_m] = np.nanAF_asnv[~mask_y] = np.nanelif bootstrap is False: # Make empty mask to skip bootstrapping.mask_y = np.zeros(len(years)).astype(bool)mask_m = np.zeros(len(t_m)).astype(bool)mask_y[:] = Truemask_m[edge:-edge] = True# Calculate trend in Airborne fraction time series.if trendcalc == 'abs': # Calculate absolute trend.AF_a_reg = slopest(t_y[mask_y], AF_a[mask_y], method=trendmethod)AF_ms_reg = slopest(t_m[mask_m], AF_ms[mask_m], method=trendmethod)AF_msnv_reg = slopest(t_m[mask_m], AF_msnv[mask_m], method=trendmethod)AF_asnv_reg = slopest(t_y[mask_y], AF_asnv[mask_y], method=trendmethod)elif trendcalc == 'rgr': # Calculate relative growth rate.AF_a_reg = slopest(t_y[mask_y], AF_a[mask_y] / float(np.mean(AF_a[mask_y])), method=trendmethod)AF_ms_reg = slopest(t_m[mask_m], AF_ms[mask_m] / np.mean(AF_ms[mask_m]), method=trendmethod)AF_msnv_reg = slopest(t_m[mask_m], AF_msnv[mask_m] / np.mean(AF_msnv[mask_m]), method=trendmethod)AF_asnv_reg = slopest(t_y[mask_y], AF_asnv[mask_y] / float(np.mean(AF_asnv[mask_y])), method=trendmethod)# Store regression results. Format: [slope, intercept, p-value, standard error]MC_a[:,mc] = [AF_a_reg[0]*uconv, AF_a_reg[1]*uconv, AF_a_reg[3]*1, AF_a_reg[4]*uconv]MC_ms[:,mc] = [AF_ms_reg[0]*uconv, AF_ms_reg[1]*uconv, AF_ms_reg[3]*1, AF_ms_reg[4]*uconv]MC_msnv[:,mc] = [AF_msnv_reg[0]*uconv, AF_msnv_reg[1]*uconv, AF_msnv_reg[3]*1, AF_msnv_reg[4]*uconv]MC_asnv[:,mc] = [AF_asnv_reg[0]*uconv, AF_asnv_reg[1]*uconv, AF_asnv_reg[3]*1, AF_asnv_reg[4]*uconv]MC_slope = np.array([MC_a[0], MC_ms[0], MC_msnv[0], MC_asnv[0]])MC_std = np.array([MC_a[3], MC_ms[3], MC_msnv[3], MC_asnv[3]])MC_p = np.array([MC_a[2], MC_ms[2], MC_msnv[2], MC_asnv[2]])# Calculate summary statistics over Monte Carlo iterations.MC_slope_median[:,i,j] = np.median(MC_slope, axis=1) # median of slopeMC_slope_std[:,i,j] = np.std(MC_slope, axis=1) # std of slopeMC_std_mean[:,i,j] = np.mean(MC_std, axis=1) # mean of stderrMC_p_mean[:,i,j] = np.mean(MC_p, axis=1) # mean of p-valuetime1 = timer.time() - time0print('Done, duration = ' + str(time1))print('Monte carlo Finished')if filename is not None:np.savez(filename, MC_slope_median=MC_slope_median, MC_slope_std=MC_slope_std, MC_std_mean=MC_std_mean, MC_p_mean=MC_p_mean)return MC_slope_median, MC_slope_std, MC_std_mean, MC_p_meanMC = 1000
size = 50
MCarr = calc_AF_MonteCarlo_ARR(MC, size, filename=wdir + 'Marle_et_al_Nature_AirborneFraction_MC%s_MK_run%sx%s_TREND%s' % (MC, size, size, trendcalc))LINgcp = np.array(MCarr)[:,:,size+1,size+1] # result for linear fit of GCP time series.
LINhan = np.array(MCarr)[:,:,size+2,size+2] # result for linear fit of Hougthon time series.
LINnew = np.array(MCarr)[:,:,size+3,size+3] # result for linear fit of This study time series.
ARR = np.array(MCarr)[:,:,:size+1,:size+1] # result array for range of intercepts and slopes.def plot_Figure4(fign, filename=None):''' Plot paper Figure 4:param fign: figure number:param filename: filename of saved figure (.pdf format). If no filename, figure is only displayed and not saved.:return: Figure 4'''fontsize = 7fontsize_legend = 5.5if trendcalc == 'rgr':clevels = np.linspace(-0.5,0.5,11)elif trendcalc == 'abs':clevels = np.linspace(-0.02,0.02,11)MC_slope_median = ARR[0]MC_slope_std = ARR[1]MC_std_mean = ARR[2]MC_p_mean = ARR[3]a = np.linspace(-0.005, 0.025, size + 1)m = np.linspace(0.8, 1.5 + (1 / 100.), size + 1)mesha = np.zeros((size+1, size+1))meshm = np.zeros((size+1, size+1))for i in range(size+1):for j in range(size+1):mesha[i,j] = a[i]meshm[i,j] = m[j]LUgcp_slope, _ = np.polyfit(t_y, LUgcp, 1) # determine slope and b for lu datasetsLUhan_slope, _ = np.polyfit(t_y, LUhan, 1)LUnew_slope, _ = np.polyfit(t_y, LUnew, 1)LUgcp_mean = np.mean(LUgcp) # determine mean for lu datasetsLUhan_mean = np.mean(LUhan)LUnew_mean = np.mean(LUnew)# Create figurefig = plt.figure(fign, figsize=(89*mm,89*0.75*mm))ax = fig.add_subplot(111)d = size/2.5 # For visualization; reducing the amount of corner points smoothens the lines.d = size/1.5mesha_plot = scipy.ndimage.zoom(mesha, 1/d)meshm_plot = scipy.ndimage.zoom(meshm, 1/d)data_plot = scipy.ndimage.zoom(MC_slope_median[iset][:size+1,:size+1], 1/d)p = ax.contourf(mesha_plot, meshm_plot, data_plot, levels=clevels, vmin=clevels[0], vmax=clevels[-1], cmap=cmap)c = ax.contour(mesha_plot, meshm_plot, data_plot, colors='black', linestyles='--', alpha=0.0) # Dummy plot to retrieve linezero_line = c.collections[4].get_paths()[0].verticesfitline_zero = np.poly1d(np.polyfit(zero_line[:,0], zero_line[:,1] ,1))ax.plot(a, fitline_zero(a), c='black', linewidth=1.2)slope_std_0 = np.mean(MC_slope_std[iset][(MC_slope_median[iset] > -0.001) & (MC_slope_median[iset] < 0.001)])l_ERR_upper = (MC_slope_median[iset] > slope_std_0).astype(int)l_ERR_lower = (MC_slope_median[iset] < -slope_std_0).astype(int)l_ERR_upper_arr = scipy.ndimage.interpolation.zoom(l_ERR_upper.astype(float), 2, order=1)l_ERR_lower_arr = scipy.ndimage.interpolation.zoom(l_ERR_lower.astype(float), 2, order=1)l_ERR_upper_arr = np.reshape(l_ERR_upper_arr, (51,2,51,2)).mean(1).mean(2)l_ERR_lower_arr = np.reshape(l_ERR_lower_arr, (51,2,51,2)).mean(1).mean(2)l_ERR_upper_arr = np.isclose(l_ERR_upper_arr, np.full((51, 51), 0.5), atol=0.45)l_ERR_lower_arr = np.isclose(l_ERR_lower_arr, np.full((51, 51), 0.5), atol=0.45)ERRupper_fit = np.poly1d(np.polyfit(mesha[l_ERR_upper_arr], meshm[l_ERR_upper_arr], 1))ERRlower_fit = np.poly1d(np.polyfit(mesha[l_ERR_lower_arr], meshm[l_ERR_lower_arr], 1))ax.plot(a, ERRupper_fit(a), color='black', linestyle='--', alpha=0.8, linewidth=1.2)ax.plot(a, ERRlower_fit(a), color='black', linestyle='--', alpha=0.8, linewidth=1.2)#ax.scatter([LUgcp_slope, LUhan_slope, LUnew_slope], [LUgcp_mean, LUhan_mean, LUnew_mean], color='black', s=50)ax.scatter([LUgcp_slope], [LUgcp_mean], color=Cgcp, s=25, edgecolors='black', linewidth=1.0, zorder=100)ax.scatter([LUhan_slope], [LUhan_mean], color=Chan, s=25, edgecolors='black', linewidth=1.0, zorder=100)ax.scatter([LUnew_slope], [LUnew_mean], color=Cnew, s=25, edgecolors='black', linewidth=1.0, zorder=100)# Set axes#ax.xaxis.set_tick_params(pad=10)ax.set_xlim(np.min(mesha), np.max(mesha)) #-0.001)ax.set_ylim(np.min(meshm), 1.5) #np.max(meshm))ax.set_adjustable('box')ax.grid(True)ax.tick_params(axis='both', which='major', labelsize=fontsize)ax.set_xlabel('Slope of LULCC emissions (Pg C yr$^{-2}$)', fontsize=fontsize)ax.set_ylabel('Average of LULCC emissions (Pg C yr$^{-1}$)', fontsize=fontsize)if trendcalc == 'rgr':cb = fig.colorbar(p, ticks=clevels)cb.set_label('Relative growth rate, RGR (% yr$^{-1}$)', size=fontsize)elif trendcalc == 'abs':cb = fig.colorbar(p, ticks=clevels)cb.set_label('Trend (decade$^{-1}$)', size=fontsize)ax.set_xticks([-0.005,0.005,0.015,0.025])cb.ax.tick_params(labelsize=fontsize)ax.xaxis.set_minor_locator(MultipleLocator(0.005))# Add scatter labelst1 = ax.text(LUgcp_slope+0.001, LUgcp_mean, u'GCP, Friedlingstein et al. (2020)', fontsize=fontsize_legend)t2 = ax.text(LUhan_slope+0.009, LUhan_mean+0.025, 'Houghton & Nassikas (2017)', verticalalignment='bottom', horizontalalignment='right', fontsize=fontsize_legend)t3 = ax.text(LUnew_slope-0.001, LUnew_mean, 'This study', verticalalignment='top', horizontalalignment='right', fontsize=fontsize_legend)t1.set_bbox(dict(facecolor='white', alpha=1.0, edgecolor='grey', linewidth=0.9, pad=2.0))t2.set_bbox(dict(facecolor='white', alpha=1.0, edgecolor='grey', linewidth=0.9, pad=2.0))t3.set_bbox(dict(facecolor='white', alpha=1.0, edgecolor='grey', linewidth=0.9, pad=2.0))plt.tight_layout()fig.subplots_adjust(left=0.14, right=0.88)if filename:plt.savefig(filename + '.pdf', bbox_inches='tight')else:plt.show()
plot_Figure4(6, filename=wdir + 'Marle_et_al_Nature_AirborneFraction_Figure4_MC%s' % MC)print('End of script.')
相关文章:

其他:Python语言绘图合集
文章目录 介绍注意导入数据函数模块画图 介绍 python语言的科研绘图合集 注意 This dataset includes the following (All files are preceded by "Marle_et_al_Nature_AirborneFraction_"):- "Datasheet.xlsx": Excel dataset containing all annual a…...

处理 Vue3 中隐藏元素刷新闪烁问题
一、问题说明 页面刷新,原本隐藏的元素会一闪而过。 效果展示: 页面的导航栏通过路由跳转中携带的 meta 参数控制导航栏的 显示/隐藏,但在实践过程中发现,虽然元素隐藏了,但是刷新页面会出现闪烁的问题。 项目源码&…...

【MySQL】数据目录迁移
一、使用场景 使用该方法一般是数据目录所在磁盘不支持扩展,只能通过新加磁盘来扩展数据目录磁盘空间。通常是Windows服务器,或者是Linux服务器的mysql数据目录的磁盘没有使用lvm。 二、准备工作 1. 新磁盘初始化,达到可使用状态 2. 需要自己…...

【项目安全设计】软件系统安全设计规范和标准(doc原件)
1.1安全建设原则 1.2 安全管理体系 1.3 安全管理规范 1.4 数据安全保障措施 1.4.1 数据库安全保障 1.4.2 操作系统安全保障 1.4.3 病毒防治 1.5安全保障措施 1.5.1实名认证保障 1.5.2 接口安全保障 1.5.3 加密传输保障 1.5.4终端安全保障 资料获取:私信或者进主页。…...

INS淡绿色风格人像街拍Lr调色教程,手机滤镜PS+Lightroom预设下载!
调色介绍 INS 淡绿色风格人像街拍通过 Lightroom 调色可以营造出清新、自然、时尚的视觉效果。这种风格以淡绿色为主色调,给人一种宁静、舒适的感觉。 预设信息 调色风格:INS风格预设适合类型:人像,街拍,自拍&#…...

python 实现最小路径和算法
最小路径和算法介绍 最小路径和问题通常指的是在一个网格(如二维数组)中,找到从起点(如左上角)到终点(如右下角)的一条路径,使得路径上经过的元素值之和最小。这类问题可以通过多种…...

Vue3实现动态菜单功能
文章目录 0.效果演示1.搭建Vue3项目1.1 vite 脚手架创建 Vue3 项目1.2 设置文件别名1.3 安装配置 element-plus1.4 安装配置路由2.登录页面3.后台管理页面3.1 搭建后台框架3.2 左侧菜单栏3.3 header 用户信息3.4 主要内容3.5 footer4.配置静态路由5.记录激活菜单5.1 el-menu 绑…...

Qt+VS2019+大恒相机相机回调方式总结
一、前言 大恒驱动安装完成后,在安装目录有SDK调用文档,里面有更详细的调用介绍,此文档对近期做的Demo做一个回顾性总结。 二、调用流程概述 三、针对性内容介绍: 1. 在执行相机操作之前,需要先执行此代码࿱…...

Python库pandas之六
Python库pandas之六 输入/输出read_sql函数应用实列 输入/输出 read_sql 函数 词法:pandas.read_sql(sql, con, index_colNone, coerce_floatTrue, paramsNone, parse_datesNone, columnsNone, chunksizeNone, dtype_backend<no_default>, dtypeNone) rea…...
[C++]使用纯opencv部署yolov11-seg实例分割onnx模型
【算法介绍】 在C中使用纯OpenCV部署YOLOv11-seg进行实例分割是一项具有挑战性的任务,因为YOLOv11通常是用PyTorch等深度学习框架实现的,而OpenCV本身并不直接支持加载和运行PyTorch模型。然而,可以通过一些间接的方法来实现这一目标&#x…...

PAT甲级-1122 Hamiltonian Cycle
题目 题目大意 给定一个图和几组顶点,判断每组顶点是否能构成一个哈密顿回路。 知识点 哈密顿回路满足几点要求:构成一个封闭环,并且经过所有顶点,每个顶点经过一次。 即满足第一个顶点值和最后一个顶点值相等;只有…...

Java 插入排序
插入排序(Insertion Sort)是一种简单直观的排序算法。它的工作原理是通过构建有序序列,对于未排序数据,在已排序序列中从后向前扫描,找到相应位置并插入。以下是插入排序的Java实现: public class Inserti…...

随机掉落的项目足迹:Vue3中vite.config.ts配置代理服务器解决跨域问题
跨域问题产生的原因:浏览器同源策略 后面的通俗解释小标题下的内容是便于大家理解同源策略和跨域问题。 而同源策略和跨域问题这两个小标题下的内容虽然比较专业不容易阅读,但是还是建议大家花时间理解并记忆,因为这是前端面试中的常考点。…...

C++笔记之标准库和boost库中bind占位符_1的写法差异
C++笔记之标准库和boost库中bind占位符_1的写法差异 code review! 参考博文: C++新特性探究(十五):bind 在C++中,_1 和 std::placeholders::_1 都用于表示占位符,但它们有不同的上下文:...

二分查找
文章目录 1.算法思想2.代码实现(1)循环实现(2)递归实现 3.题目练习 1.算法思想 二分查找(折半查找):有序数组(升序或降序,可以不连续),每次缩小一半的区间。 时间复杂度:O(log n) 空间复杂度:循环实现是 O(1)…...

关注、取关、Redis实现共同关注、 博客推送与分页查询
Resourceprivate StringRedisTemplate stringRedisTemplate;Resourceprivate IUserService userService;Overridepublic Result follow(Long followUserId, Boolean isFollow) {//1.获取登陆的用户Long userId UserHolder.getUser().getId();//1.判断是关注还是取关if(isFollo…...

专业高清录屏软件!Mirillis Action v4.40 解锁版下载,小白看了都会的安装方法
Mirillis Action!(暗神屏幕录制软件)专业高清屏幕录像软件,被誉为游戏视频三大神器之一。这款屏幕录制软件和游戏录制软件,拥有三大硬件加速技术,支持以超高清视频画质录制桌面和实况直播,超清视频画质&…...

胤娲科技:AI重塑会议——灵动未来,会议新纪元
你是否曾经历过这样的会议场景:会议纪要不准确,人名张冠李戴;错过会议,却无从回顾关键内容;会议效率低下,时间白白流逝? 这些问题仿佛成了现代会议的“顽疾”。然而,随着AI技术的飞速…...

Python画笔案例-080 绘制 颜色亮度测试
1、绘制 颜色亮度测试 通过 python 的turtle 库绘制 颜色亮度测试,如下图: 2、实现代码 绘制 颜色亮度测试,以下为实现代码: """颜色亮度测试.py本程序需要coloradd模块支持,请在cmd窗口,即命令提示符下输入pip install coloradd进行安装。本程序演示brig…...

MATLAB工具库:数据统计分析工具MvCAT、MhAST等
MATLAB工具库:数据统计分析工具MvCAT、MhAST等 工具1:Multivariate Copula Analysis Toolbox (MvCAT)MATLAB中运行 工具2:Multi-hazard Scenario Analysis Toolbox (MhAST) 参考 The University of California-软件库-Software 工具1…...

Android Wi-Fi 连接失败日志分析
1. Android wifi 关键日志总结 (1) Wi-Fi 断开 (CTRL-EVENT-DISCONNECTED reason3) 日志相关部分: 06-05 10:48:40.987 943 943 I wpa_supplicant: wlan0: CTRL-EVENT-DISCONNECTED bssid44:9b:c1:57:a8:90 reason3 locally_generated1解析: CTR…...

日语AI面试高效通关秘籍:专业解读与青柚面试智能助攻
在如今就业市场竞争日益激烈的背景下,越来越多的求职者将目光投向了日本及中日双语岗位。但是,一场日语面试往往让许多人感到步履维艰。你是否也曾因为面试官抛出的“刁钻问题”而心生畏惧?面对生疏的日语交流环境,即便提前恶补了…...

SciencePlots——绘制论文中的图片
文章目录 安装一、风格二、1 资源 安装 # 安装最新版 pip install githttps://github.com/garrettj403/SciencePlots.git# 安装稳定版 pip install SciencePlots一、风格 简单好用的深度学习论文绘图专用工具包–Science Plot 二、 1 资源 论文绘图神器来了:一行…...
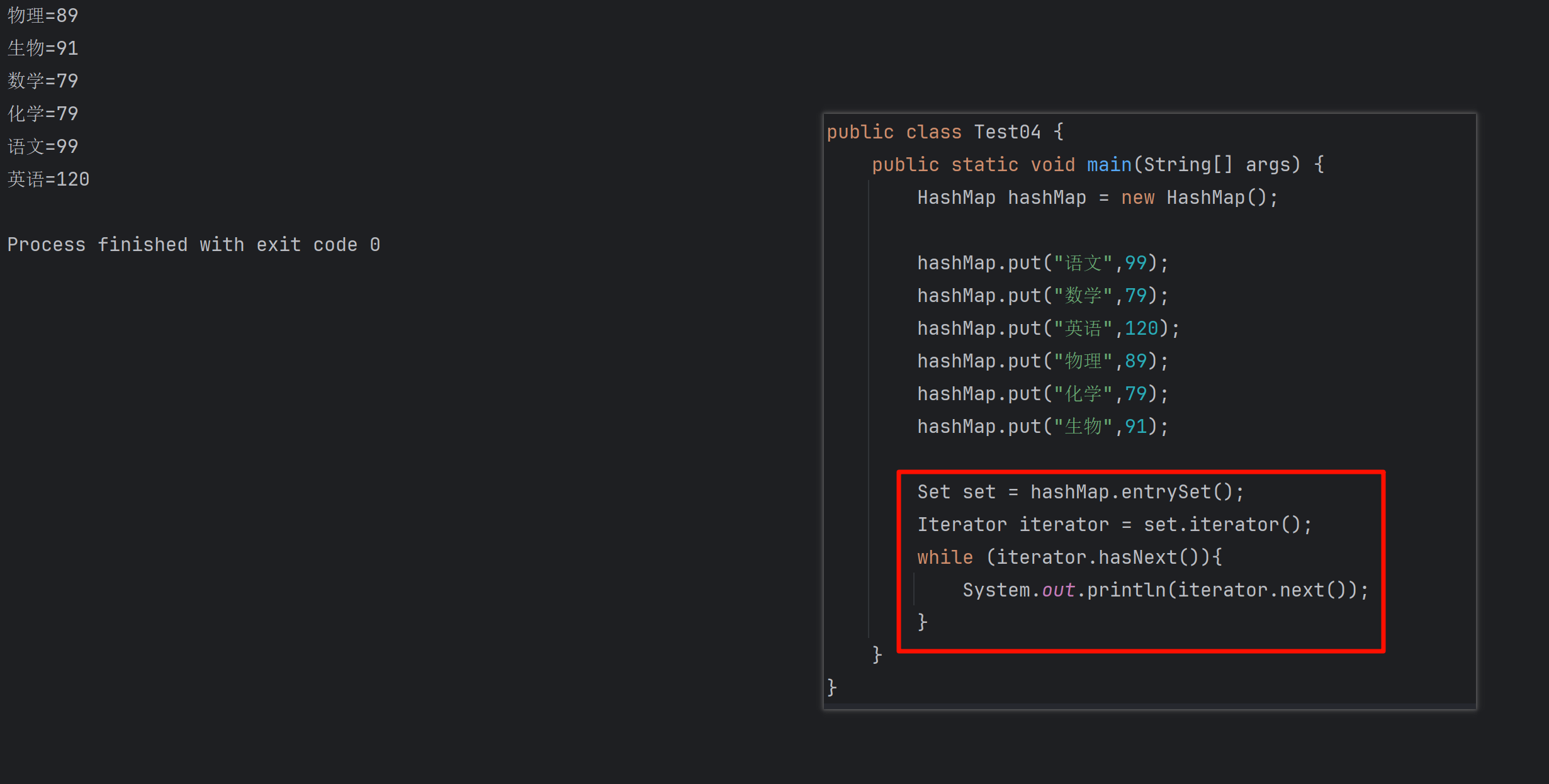
遍历 Map 类型集合的方法汇总
1 方法一 先用方法 keySet() 获取集合中的所有键。再通过 gey(key) 方法用对应键获取值 import java.util.HashMap; import java.util.Set;public class Test {public static void main(String[] args) {HashMap hashMap new HashMap();hashMap.put("语文",99);has…...

使用van-uploader 的UI组件,结合vue2如何实现图片上传组件的封装
以下是基于 vant-ui(适配 Vue2 版本 )实现截图中照片上传预览、删除功能,并封装成可复用组件的完整代码,包含样式和逻辑实现,可直接在 Vue2 项目中使用: 1. 封装的图片上传组件 ImageUploader.vue <te…...
零基础设计模式——行为型模式 - 责任链模式
第四部分:行为型模式 - 责任链模式 (Chain of Responsibility Pattern) 欢迎来到行为型模式的学习!行为型模式关注对象之间的职责分配、算法封装和对象间的交互。我们将学习的第一个行为型模式是责任链模式。 核心思想:使多个对象都有机会处…...

3403. 从盒子中找出字典序最大的字符串 I
3403. 从盒子中找出字典序最大的字符串 I 题目链接:3403. 从盒子中找出字典序最大的字符串 I 代码如下: class Solution { public:string answerString(string word, int numFriends) {if (numFriends 1) {return word;}string res;for (int i 0;i &…...

什么?连接服务器也能可视化显示界面?:基于X11 Forwarding + CentOS + MobaXterm实战指南
文章目录 什么是X11?环境准备实战步骤1️⃣ 服务器端配置(CentOS)2️⃣ 客户端配置(MobaXterm)3️⃣ 验证X11 Forwarding4️⃣ 运行自定义GUI程序(Python示例)5️⃣ 成功效果
基于matlab策略迭代和值迭代法的动态规划
经典的基于策略迭代和值迭代法的动态规划matlab代码,实现机器人的最优运输 Dynamic-Programming-master/Environment.pdf , 104724 Dynamic-Programming-master/README.md , 506 Dynamic-Programming-master/generalizedPolicyIteration.m , 1970 Dynamic-Programm…...
:工厂方法模式、单例模式和生成器模式)
上位机开发过程中的设计模式体会(1):工厂方法模式、单例模式和生成器模式
简介 在我的 QT/C 开发工作中,合理运用设计模式极大地提高了代码的可维护性和可扩展性。本文将分享我在实际项目中应用的三种创造型模式:工厂方法模式、单例模式和生成器模式。 1. 工厂模式 (Factory Pattern) 应用场景 在我的 QT 项目中曾经有一个需…...
