CellMarker | 人骨骼肌组织细胞Marker大全!~(强烈建议火速收藏!)
1写在前面
分享一下最近看到的2篇paper关于骨骼肌组织的细胞Marker,绝对的Atlas级好东西。👍
希望做单细胞的小伙伴觉得有用哦。😏
2常用marker(一)
general_mrkrs <- c(
'MYH7', 'TNNT1', 'TNNT3', 'MYH1', 'MYH2', "CKM", "MB", # Myofibers
'PAX7', 'DLK1', # MuSCs
'PDGFRA', 'DCN', 'ANGPTL7', 'OSR2', 'NGFR', 'SLC22A3','ITGA6', # Fibroblasts
'FMOD', 'TNMD' , 'MKX', # Tenocytes
'MPZ', 'MBP', # Schwann cells
'CDH2', 'L1CAM', # SCG
'MSLN', 'ITLN1', # mesothelium
"ADIPOQ", "PLIN1", # adipocytes
'PTPRC', 'CD3D', 'IL7R', # T cells
'NKG7', 'PRF1', #NK cells
'CD79A', "TCL1A", # B cells
'MZB1', 'JCHAIN', # B plasma
"CD14", "FCGR3A",'S100A8', 'S100A12', # Mono
"CD163", "C1QA", # Macrop
"XCR1", "CLEC9A", # cDC1 "CADM1",
"CD1C", "CLEC10A", "CCR7", # cDC2
'LILRA4', 'IL3RA', "IRF7", # pDC
'FCGR3B', 'CSF3R', 'SORL1', # Neutrophils
'EPX', 'PRG2', # Eosinophils 'CLC'
'TPSB2', 'MS4A2', # Mast cells
'PECAM1', 'HEY1','CLU', # art EC
'CA4', 'LPL', # capEC
'ACKR1', 'SELE', # venEC
'LYVE1', 'TFF3', # lymphEC
'RGS5','ABCC9', # pericytes
'MYH11', 'ACTA2', # SMC
'HBA1', #RBC
)
出自下面paper:👇
Human skeletal muscle aging atlas. Veronika R. Kedlian, Yaning Wang, Tianliang Liu, Xiaoping Chen, Liam Bolt, Catherine Tudor, Zhuojian Shen, Eirini S. Fasouli, Elena Prigmore, Vitalii Kleshchevnikov, Jan Patrick Pett, Tong Li, John E G Lawrence, Shani Perera, Martin Prete, Ni Huang, Qin Guo, Xinrui Zeng, Lu Yang, Krzysztof Polański, Nana-Jane Chipampe, Monika Dabrowska, Xiaobo Li, Omer Ali Bayraktar, Minal Patel, Natsuhiko Kumasaka, Krishnaa T. Mahbubani, Andy Peng Xiang, Kerstin B. Meyer, Kourosh Saeb-Parsy, Sarah A Teichmann & Hongbo Zhang 2024 Apr.
3常用marker(二)
Mural Cell Markers
#SMOOTH MUSCLE CELLS
FeaturePlot(df.harmony, features = "MYH11", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ACTA2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TAGLN", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#PERICYTES
FeaturePlot(df.harmony, features = "RGS5", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CSPG4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PDGFRB", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Glial Cells Markers
FeaturePlot(df.harmony, features = "PROX1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MPZ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "NCAM1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CDH19", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SOX10", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PLP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Adipocites Markers
FeaturePlot(df.harmony, features = "PLIN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ADIPOQ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MMRN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CCL21", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Tenocytes Markers
FeaturePlot(df.harmony, features = "FMOD", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TNMD", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL22A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SCX", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "DLG2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "FBN1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Endothelial Markers
#FeaturePlot(df.harmony, features = "PCDHA6", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#ARTERIAL
FeaturePlot(df.harmony, features = "FBLN5", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "DLL4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "SEMA3G", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#CAPILLARIES
FeaturePlot(df.harmony, features = "RGCC", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#VENOUS
FeaturePlot(df.harmony, features = "EPHB4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
Myonuclei Markers
FeaturePlot(df.harmony, features = "TTN", min.cutoff = "q9", order = T, cols = c("lightblue", "navy"), raster = FALSE)
#IMMATURE MYOCYTE
FeaturePlot(df.harmony, features = "MYMX", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYOG", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
#REG MYONUCLEI
FeaturePlot(df.harmony, features = "FLNC", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYH3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "MYH8", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "XIRP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
NMJ Myonuclei Markers (Neuromuscular junction)
#NMJ
FeaturePlot(df.harmony, features = "CHRNE", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "CHRNA1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PRKAR1A", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL25A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "UTRN", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COLQ", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "ABLIM2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "VAV3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "UFSP1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
MTJ Myonuclei Markers (Myotendinous junction)
#MTJ
FeaturePlot(df.harmony, features = "COL22A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "PIEZO2", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL24A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL6A1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "FSTL1", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "COL6A3", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(df.harmony, features = "TIGD4", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
FeaturePlot(mini_df.harmony.harmony, features = "EYS", min.cutoff = "q9", order = TRUE, cols = c("lightblue", "navy"), raster = FALSE)
出自下面paper:👇
Lai, Y., Ramírez-Pardo, I., Isern, J. et al. Multimodal cell atlas of the ageing human skeletal muscle. Nature (2024).
点个在看吧各位~ ✐.ɴɪᴄᴇ ᴅᴀʏ 〰
📍 🤩 LASSO | 不来看看怎么美化你的LASSO结果吗!?
📍 🤣 chatPDF | 别再自己读文献了!让chatGPT来帮你读吧!~
📍 🤩 WGCNA | 值得你深入学习的生信分析方法!~
📍 🤩 ComplexHeatmap | 颜狗写的高颜值热图代码!
📍 🤥 ComplexHeatmap | 你的热图注释还挤在一起看不清吗!?
📍 🤨 Google | 谷歌翻译崩了我们怎么办!?(附完美解决方案)
📍 🤩 scRNA-seq | 吐血整理的单细胞入门教程
📍 🤣 NetworkD3 | 让我们一起画个动态的桑基图吧~
📍 🤩 RColorBrewer | 再多的配色也能轻松搞定!~
📍 🧐 rms | 批量完成你的线性回归
📍 🤩 CMplot | 完美复刻Nature上的曼哈顿图
📍 🤠 Network | 高颜值动态网络可视化工具
📍 🤗 boxjitter | 完美复刻Nature上的高颜值统计图
📍 🤫 linkET | 完美解决ggcor安装失败方案(附教程)
📍 ......




本文由 mdnice 多平台发布
相关文章:

CellMarker | 人骨骼肌组织细胞Marker大全!~(强烈建议火速收藏!)
1写在前面 分享一下最近看到的2篇paper关于骨骼肌组织的细胞Marker,绝对的Atlas级好东西。👍 希望做单细胞的小伙伴觉得有用哦。😏 2常用marker(一) general_mrkrs <- c( MYH7, TNNT1, TNNT3, MYH1, MYH2, "C…...

游戏名台词大赏
文章目录 原神(圈内) 崩坏:星穹铁道(圈内) 崩坏3(圈内) 原神 只要不失去你的崇高,整个世界都会为你敞开。 总会有地上的生灵,敢于直面雷霆的威光。 谁也没有见过风&…...

OpenCV如何在图像中寻找轮廓(60)
返回:OpenCV系列文章目录(持续更新中......) 上一篇:OpenCV如何模板匹配(59) 下一篇 :OpenCV检测凸包(61) 目标 在本教程中,您将学习如何: 使用 OpenCV 函数 cv::findContours使用 OpenCV 函数 cv::d rawContours …...

java 泛型题目讲解
泛型的知识点 泛型仅存在于编译时期,编译期间JAVA将会使用Object类型代替泛型类型,在运行时期不存在泛型;且所有泛型实例共享一个泛型类 public class Main{public static void main(String[] args){ArrayList<String> list1new Arra…...
)
pptx 文件版面分析-- python-pptx(python 文档解析提取)
安装 pip install python-pptx -i https://pypi.tuna.tsinghua.edu.cn/simple --ignore-installedpptx 解析代码实现 from pptx import Presentation file_name "rag_pptx/test1.pptx" # 打开.pptx文件 ppt Presentation(file_name) for slide in ppt.slides:#pr…...

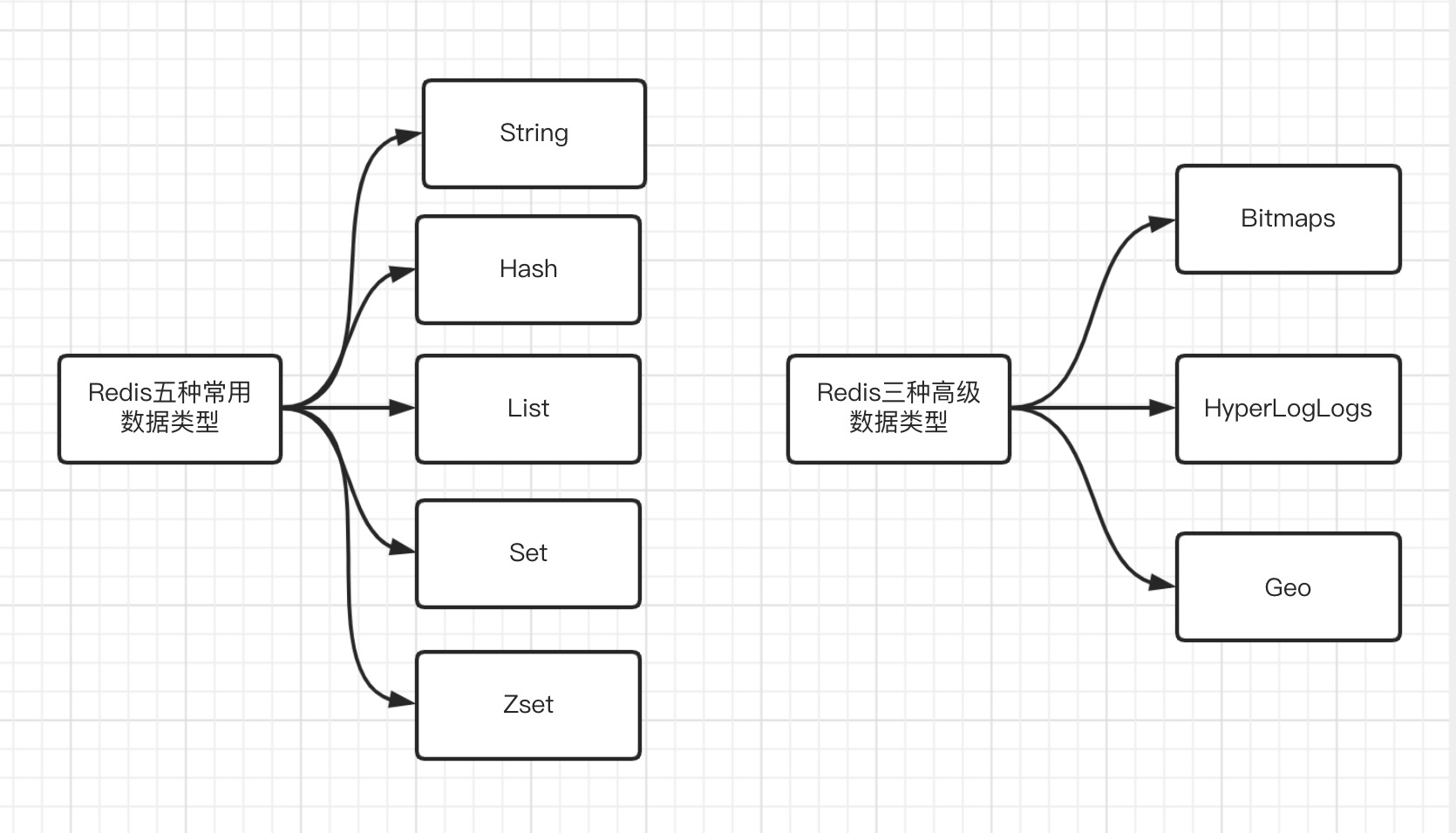
http的basic 认证方式
写在前面 本文看下http的basic auth认证方式。 1:什么是basic auth认证 basic auth是一种http协议规范中的一种认证方式,即一种证明你就是你的方式。更进一步的它是一种规范,这种规范是这样子,如果是服务端使用了basic auth认证…...

【信息系统项目管理师练习题】信息系统治理
IT治理的核心是关注以下哪项内容? a) 人员培训和发展计划 b) IT定位和信息化建设与数字化转型的责权利划分 c) 业务流程的绩效管理 d) IT基础设施的优化利用 答案: b) IT定位和信息化建设与数字化转型的责权利划分 IT治理体系框架的组成部分包括以下哪些? a) IT战略目标、IT治…...

RabbitMQ之顺序消费
什么是顺序消费 例如:业务上产生者发送三条消息, 分别是对同一条数据的增加、修改、删除操作, 如果没有保证顺序消费,执行顺序可能变成删除、修改、增加,这就乱了。 如何保证顺序性 一般我们讨论如何保证消息的顺序性&…...

轻松上手的LangChain学习说明书
一、Langchain是什么? 如今各类AI模型层出不穷,百花齐放,大佬们开发的速度永远遥遥领先于学习者的学习速度。。为了解放生产力,不让应用层开发人员受限于各语言模型的生产部署中…LangChain横空出世界。 Langchain可以说是现阶段…...

【论文笔记】Training language models to follow instructions with human feedback A部分
Training language models to follow instructions with human feedback A 部分 回顾一下第一代 GPT-1 : 设计思路是 “海量无标记文本进行无监督预训练少量有标签文本有监督微调” 范式;模型架构是基于 Transformer 的叠加解码器(掩码自注意…...

嵌入式交叉编译:x265
下载 multicoreware / x265_git / Downloads — Bitbucket 解压编译 BUILD_DIR${HOME}/build_libs CROSS_NAMEaarch64-mix210-linuxcd build/aarch64-linuxmake cleancmake \-G "Unix Makefiles" \-DCMAKE_C_COMPILER${CROSS_NAME}-gcc \-DCMAKE_CXX_COMPILER${CR…...
一、Redis五种常用数据类型
Redis优势: 1、性能高—基于内存实现数据的存储 2、丰富的数据类型 5种常用,3种高级 3、原子—redis的所有单个操作都是原子性,即要么成功,要么失败。其多个操作也支持采用事务的方式实现原子性。 Redis特点: 1、支持…...

C语言动态内存管理malloc、calloc、realloc、free函数、内存泄漏、动态内存开辟的位置等的介绍
文章目录 前言一、为什么存在动态内存管理二、动态内存函数的介绍1. malloc函数2. 内存泄漏3. 动态内存开辟位置4. free函数5. calloc 函数6. realloc 函数7. realloc 传空指针 总结 前言 C语言动态内存管理malloc、calloc、realloc、free函数、内存泄漏、动态内存开辟的位置等…...

最近惊爆谷歌裁员
Python团队还没解散完,谷歌又对Flutter、Dart动手了。 什么原因呢,猜测啊。 谷歌裁员Python的具体原因可能是因为公司在进行技术栈的调整和优化。Python作为一种脚本语言,在某些情况下可能无法提供足够的性能或者扩展性,尤其是在…...

音频可视化:原生音频API为前端带来的全新可能!
音频API是一组提供给网页开发者的接口,允许他们直接在浏览器中处理音频内容。这些API使得在不依赖任何外部插件的情况下操作和控制音频成为可能。 Web Audio API 可以进行音频的播放、处理、合成以及分析等操作。借助于这些工具,开发者可以实现自定义的音…...
)
【中等】保研/考研408机试-动态规划1(01背包、完全背包、多重背包)
背包问题基本上都是模板题,重点:弄熟多重背包模板 dp[j]max(dp[j-v[i]]w[i],dp[j]) //核心思路代码(一维数组版) dp[i][j]max(dp[i-1][j], dp[i-1][j-v[i]]w[i])//二维数字版 一、 0-1背包 一般输入两个变量:体积&…...

[DEMO]给两个字符串取交集的词语
要求:2个英文字符串中,取相同的大于等于4个字母的词组 比如: 字符串1:" xingMeiLingabcdef WorldHello", 字符串2:"mnjqlup WorldLingLing xingMeiLingHello" 获取交接: [xingMeiLing…...

leetcode53-Maximum Subarray
题目 给你一个整数数组 nums ,请你找出一个具有最大和的连续子数组(子数组最少包含一个元素),返回其最大和。 子数组 是数组中的一个连续部分。 示例 1: 输入:nums [-2,1,-3,4,-1,2,1,-5,4] 输出…...

Python 基于 OpenCV 视觉图像处理实战 之 OpenCV 简单人脸检测/识别实战案例 之七 简单进行人脸检测并添加面具特效实现
Python 基于 OpenCV 视觉图像处理实战 之 OpenCV 简单人脸检测/识别实战案例 之七 简单进行人脸检测并添加面具特效实现 目录...
【go项目01_学习记录06】
学习记录 1 使用中间件1.1 测试一下1.2 push代码 2 URI 中的斜杆2.1 StrictSlash2.2 兼容 POST 请求 1 使用中间件 代码中存在重复率很高的代码 w.Header().Set("Content-Type", "text/html; charsetutf-8")统一对响应做处理的,我们可以使用中…...
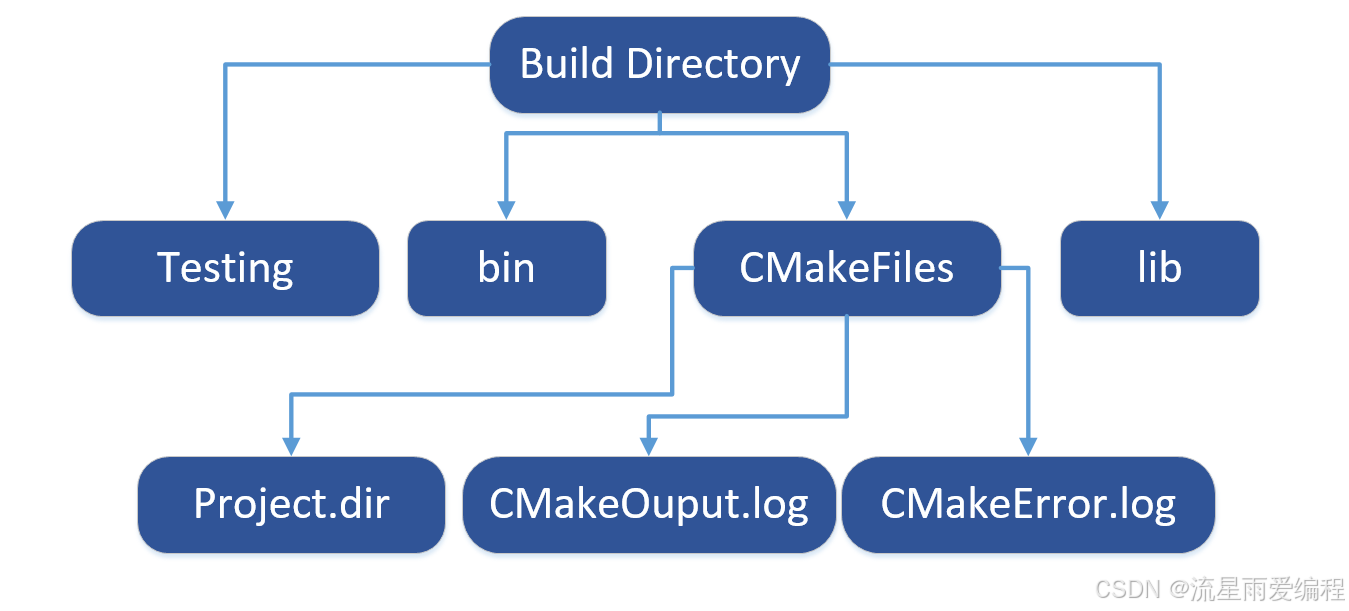
CMake基础:构建流程详解
目录 1.CMake构建过程的基本流程 2.CMake构建的具体步骤 2.1.创建构建目录 2.2.使用 CMake 生成构建文件 2.3.编译和构建 2.4.清理构建文件 2.5.重新配置和构建 3.跨平台构建示例 4.工具链与交叉编译 5.CMake构建后的项目结构解析 5.1.CMake构建后的目录结构 5.2.构…...
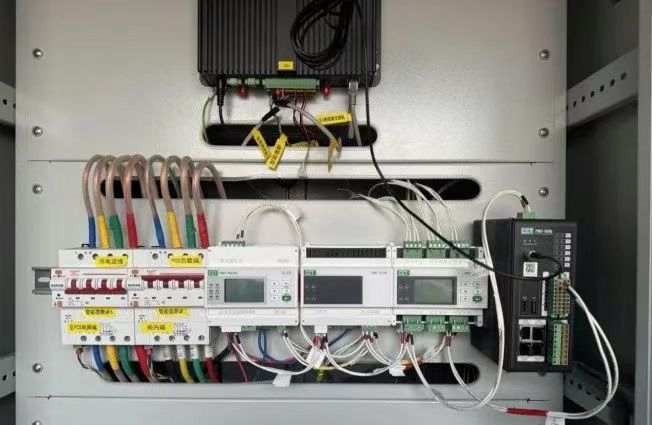
IT供电系统绝缘监测及故障定位解决方案
随着新能源的快速发展,光伏电站、储能系统及充电设备已广泛应用于现代能源网络。在光伏领域,IT供电系统凭借其持续供电性好、安全性高等优势成为光伏首选,但在长期运行中,例如老化、潮湿、隐裂、机械损伤等问题会影响光伏板绝缘层…...

爬虫基础学习day2
# 爬虫设计领域 工商:企查查、天眼查短视频:抖音、快手、西瓜 ---> 飞瓜电商:京东、淘宝、聚美优品、亚马逊 ---> 分析店铺经营决策标题、排名航空:抓取所有航空公司价格 ---> 去哪儿自媒体:采集自媒体数据进…...

Spring Cloud Gateway 中自定义验证码接口返回 404 的排查与解决
Spring Cloud Gateway 中自定义验证码接口返回 404 的排查与解决 问题背景 在一个基于 Spring Cloud Gateway WebFlux 构建的微服务项目中,新增了一个本地验证码接口 /code,使用函数式路由(RouterFunction)和 Hutool 的 Circle…...

IP如何挑?2025年海外专线IP如何购买?
你花了时间和预算买了IP,结果IP质量不佳,项目效率低下不说,还可能带来莫名的网络问题,是不是太闹心了?尤其是在面对海外专线IP时,到底怎么才能买到适合自己的呢?所以,挑IP绝对是个技…...

【Go语言基础【12】】指针:声明、取地址、解引用
文章目录 零、概述:指针 vs. 引用(类比其他语言)一、指针基础概念二、指针声明与初始化三、指针操作符1. &:取地址(拿到内存地址)2. *:解引用(拿到值) 四、空指针&am…...
代码规范和架构【立芯理论一】(2025.06.08)
1、代码规范的目标 代码简洁精炼、美观,可持续性好高效率高复用,可移植性好高内聚,低耦合没有冗余规范性,代码有规可循,可以看出自己当时的思考过程特殊排版,特殊语法,特殊指令,必须…...
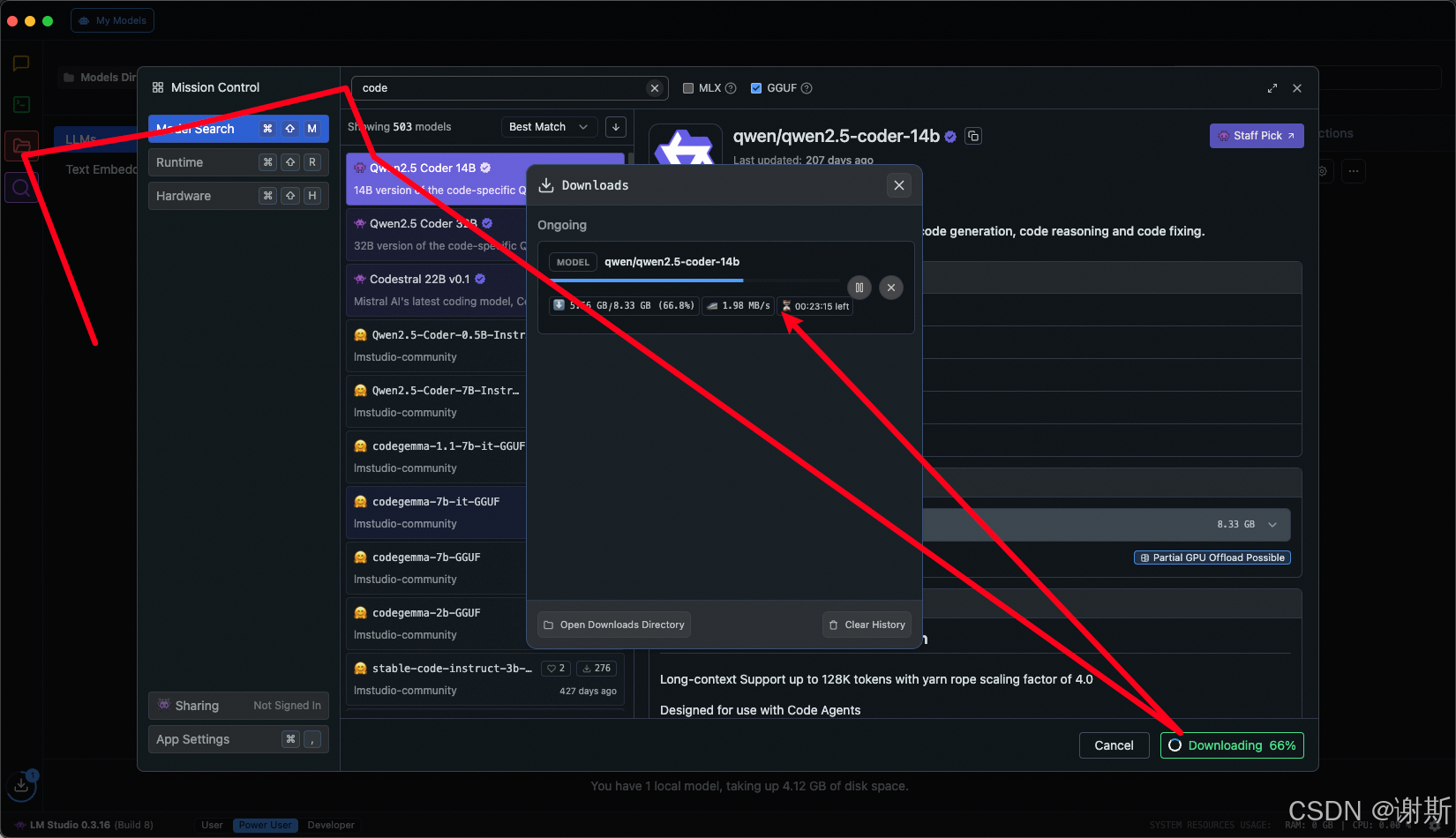
[大语言模型]在个人电脑上部署ollama 并进行管理,最后配置AI程序开发助手.
ollama官网: 下载 https://ollama.com/ 安装 查看可以使用的模型 https://ollama.com/search 例如 https://ollama.com/library/deepseek-r1/tags # deepseek-r1:7bollama pull deepseek-r1:7b改token数量为409622 16384 ollama命令说明 ollama serve #:…...

解决:Android studio 编译后报错\app\src\main\cpp\CMakeLists.txt‘ to exist
现象: android studio报错: [CXX1409] D:\GitLab\xxxxx\app.cxx\Debug\3f3w4y1i\arm64-v8a\android_gradle_build.json : expected buildFiles file ‘D:\GitLab\xxxxx\app\src\main\cpp\CMakeLists.txt’ to exist 解决: 不要动CMakeLists.…...

TSN交换机正在重构工业网络,PROFINET和EtherCAT会被取代吗?
在工业自动化持续演进的今天,通信网络的角色正变得愈发关键。 2025年6月6日,为期三天的华南国际工业博览会在深圳国际会展中心(宝安)圆满落幕。作为国内工业通信领域的技术型企业,光路科技(Fiberroad&…...
