用 Python 从零开始创建神经网络(十七):回归(Regression)
回归(Regression)
- 引言
- 1. 线性激活(Linear Activation)
- 2. 均方误差损失(Mean Squared Error Loss)
- 3. 均方误差损失导数(Mean Squared Error Loss Derivative)
- 4. 平均平方误差 (MSE) 损失代码(Mean Squared Error (MSE) Loss Code)
- 5. 平均绝对误差损失(Mean Absolute Error Loss)
- 6. 平均绝对误差损失导数(Mean Absolute Error Loss Derivative)
- 7. 平均绝对误差损失代码(Mean Absolute Error Loss Code)
- 8. 回归精度(Accuracy in Regression)
- 9. 回归模型训练(Regression Model Training)
- 到此为止的全部代码:
引言
到目前为止,我们一直在处理分类模型,目标是确定某个事物是什么。现在我们感兴趣的是基于输入来确定一个具体的数值。例如,你可能希望使用神经网络来预测明天的温度或一辆车的价格。对于这样的任务,我们需要输出更加精细的结果。这也意味着我们需要一种新的方法来衡量损失,并且需要一个新的输出层激活函数!同时,这也意味着我们所用的数据不同。我们需要包含目标标量数值的训练数据,而不是分类数据。
import matplotlib.pyplot as plt
import nnfs
from nnfs.datasets import sine_datannfs.init()X, y = sine_data()plt.plot(X, y)
plt.show()
上面的数据将生成类似的图表:

1. 线性激活(Linear Activation)
由于我们不再使用分类标签,而是要预测一个标量数值,因此我们将对输出层使用线性激活函数。该线性函数不会修改输入,而是将其直接传递到输出: y = x y = x y=x。在反向传播过程中,我们已经知道 f ( x ) = x f(x) = x f(x)=x的导数是1;因此,我们新线性激活函数的完整类定义为:
# Linear activation
class Activation_Linear:# Forward passdef forward(self, inputs):# Just remember valuesself.inputs = inputsself.output = inputs# Backward passdef backward(self, dvalues):# derivative is 1, 1 * dvalues = dvalues - the chain ruleself.dinputs = dvalues.copy()
这可能会引发一个问题:为什么我们甚至要编写一些什么都不做的代码?在前向传播中,我们只是将输入传递到输出,在反向传播中同样处理梯度,因为为了应用链式法则,我们将传入的梯度乘以导数,而导数为1。我们这样做只是为了完整性和清晰性,以便在模型定义代码中看到输出层的激活函数。从计算时间的角度来看,这几乎不会增加任何处理时间,至少不会显著影响训练时间。
现在我们只需要解决损失函数的问题!
2. 均方误差损失(Mean Squared Error Loss)
由于我们不再使用分类标签,因此无法计算交叉熵。相反,我们需要一些新的方法。回归任务中计算误差的两种主要方法是均方误差(MSE)和平均绝对误差(MAE)。
对于均方误差,我们将预测值和真实值之间的差异取平方(对于多个回归输出,每个输出都会计算差异),然后对这些平方值求平均。
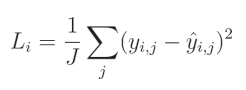
其中, y y y 表示目标值, y ^ \hat{y} y^ 表示预测值,索引 i i i 表示当前样本,索引 j j j 表示该样本中的当前输出, J J J 表示输出的数量。
这里的思想是,偏离目标值越远,惩罚越大。
3. 均方误差损失导数(Mean Squared Error Loss Derivative)
相对于预测值的平方误差偏导数为:
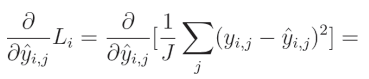
1 J \frac{1}{J} J1(输出数量 J J J 的倒数)是一个常数,可以移到导数的外部。由于我们是针对给定输出 j j j 计算导数,因此单个元素的求和结果等于该元素本身。
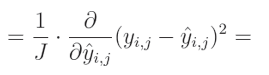
要计算一个表达式的幂的偏导数,我们需要将这个指数与该表达式相乘,然后从指数中减去1,并将其乘以内函数的偏导数:
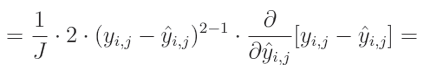
减法的偏导数等于偏导数的减法:
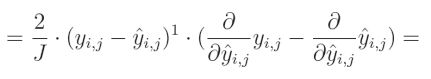
关于真实值对预测值的偏导数等于0,因为我们将其他变量视为常数。而预测值对自身的偏导数等于1,这导致结果为 0 − 1 = − 1 0-1=-1 0−1=−1。这个结果与方程的其余部分相乘,形成最终解:
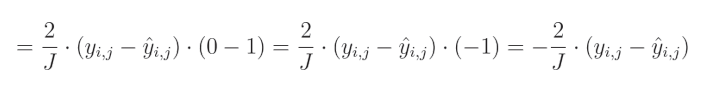
全面解决方案:
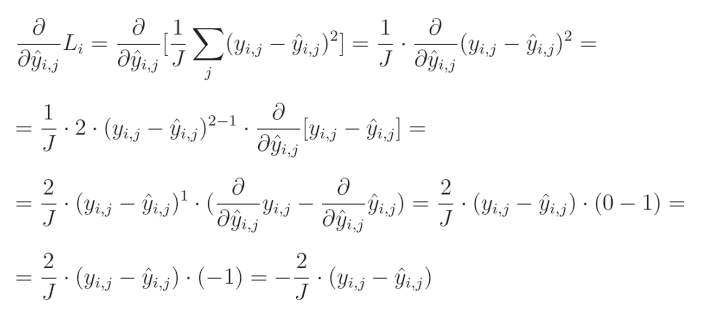
偏导数等于 − 2 -2 −2,再乘以真实值与预测值的差,然后除以输出的数量以对梯度进行归一化,从而使梯度的大小与输出的数量无关。
4. 平均平方误差 (MSE) 损失代码(Mean Squared Error (MSE) Loss Code)
MSE(均方误差)的代码实现包括了用于计算多个输出的样本损失的方程。axis=-1 与均值计算的含义在前一章节中已有详细解释,简而言之,它告诉NumPy对每个样本单独计算输出间的均值。在反向传播中,我们实现了导数方程,其结果为 − 2 -2 −2乘以真实值与预测值之间的差,并除以输出数量进行归一化。与其他损失函数实现类似,我们还对梯度按样本数量进行归一化,使其与批量大小或样本数量无关。
import numpy as np# Mean Squared Error loss
class Loss_MeanSquaredError(Loss): # L2 loss# Forward passdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean((y_true - y_pred)**2, axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Gradient on valuesself.dinputs = -2 * (y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples
5. 平均绝对误差损失(Mean Absolute Error Loss)
对于平均绝对误差(Mean Absolute Error, MAE),你需要计算单个输出中预测值与真实值之间的绝对差值,然后对这些绝对值求平均:
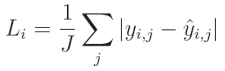
其中, y y y 表示目标值, y ^ \hat{y} y^ 表示预测值,索引 i i i 表示当前样本,索引 j j j 表示当前样本中的输出, J J J 表示输出的数量。
该函数作为损失函数时,对误差进行线性惩罚。它会产生更加稀疏的结果,并对异常值具有较强的鲁棒性,这既可能是优点,也可能是缺点。实际上,L1(MAE)损失的使用频率低于L2(MSE)损失。
6. 平均绝对误差损失导数(Mean Absolute Error Loss Derivative)
绝对误差相对于预测值的偏导数为:
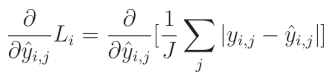
1 1 1 除以 J J J(输出数量)是一个常数,可以移到导数之外。由于我们是对给定输出 j j j 求导,所以一个元素的和等于该元素本身:
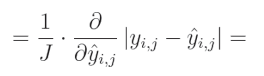
我们之前已经为L1正则化计算了绝对值的偏导数,这与L1损失类似。绝对值的导数在该值大于 0 0 0时等于 1 1 1,在该值小于 0 0 0时等于 − 1 -1 −1。当该值为 0 0 0时,导数不存在:
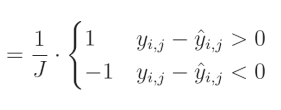
完整解决方案:
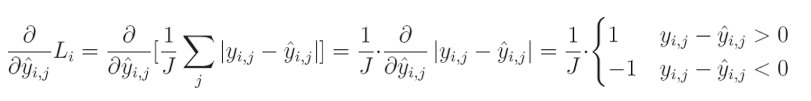
7. 平均绝对误差损失代码(Mean Absolute Error Loss Code)
均绝对误差(Mean Absolute Error, MAE)的代码与均方误差(Mean Squared Error, MSE)非常相似。前向传播中使用NumPy的np.abs()函数来计算绝对值,然后再计算均值。反向传播中,我们将使用np.sign()函数,该函数根据输入的符号返回 1 1 1或 − 1 -1 −1,如果参数等于 0 0 0,则返回 0 0 0。接着,我们将梯度根据样本数量进行归一化,使其不受批量大小或样本数量的影响:
import numpy as np# Mean Absolute Error loss
class Loss_MeanAbsoluteError(Loss): # L1 lossdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean(np.abs(y_true - y_pred), axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Calculate gradientself.dinputs = np.sign(y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples
8. 回归精度(Accuracy in Regression)
既然我们已经有了数据、激活函数和用于回归的损失计算,我们接下来要衡量模型的性能。
在使用交叉熵时,我们可以计算预测与真实目标值相等的情况数,并将其除以样本数量,从而衡量模型的准确率。然而,对于回归模型,我们面临两个问题:
- 第一个问题是,模型中的每个输出神经元(可能有很多)都是单独的输出,这与分类器不同,分类器的所有输出会共同贡献于一个预测结果。
- 第二个问题是,回归模型的预测值是浮点数,我们无法简单地检查输出值是否与真实目标值完全相等,因为它很可能不会相等——即便只有微小的差异,准确率也会被计算为 0 0 0。例如,如果你的模型预测房价,其中某个样本的目标价格是 192 , 500 192,500 192,500,而预测值是 192 , 495 192,495 192,495,那么纯粹的“是否相等”评估会返回False。但考虑到数值的量级,这样的预测在实际中可以被视为“足够接近”或正确。
对于回归任务,没有完美的方法来衡量准确率。但为了直观展示性能,最好还是有一个准确率度量。例如,流行的深度学习框架Keras会同时展示回归模型的准确率和损失,我们也将定义自己的准确率指标。
首先,我们需要一个“限制”值,我们称之为“精度”(precision)。为了计算这个精度,我们将从真实目标值中计算标准差,并将其除以 250 250 250。根据你的目标,这个值可能会有所不同。分母越大,准确率指标就会越“严格”。这里我们选择 250 250 250作为分母。以下是表示这一过程的代码:
accuracy_precision = np.std(y) / 250
然后,我们可以将这个精度值用作回归输出的“容差范围”,在比较目标值和预测值的准确性时起到缓冲作用。我们通过对真实目标值与预测值之间的差异取绝对值来进行比较。接着,我们检查这个差异是否小于我们之前计算得到的精度值:
predictions = activation2.output
accuracy = np.mean(np.absolute(predictions - y) < accuracy_precision)
9. 回归模型训练(Regression Model Training)
有了新的激活函数、损失和计算精度的方法,我们现在就可以创建模型了:
import numpy as np
import nnfs
from nnfs.datasets import spiral_data
from nnfs.datasets import sine_datannfs.init()# Dense layer
class Layer_Dense:# Layer initializationdef __init__(self, n_inputs, n_neurons,weight_regularizer_l1=0, weight_regularizer_l2=0,bias_regularizer_l1=0, bias_regularizer_l2=0):# Initialize weights and biasesself.weights = 0.01 * np.random.randn(n_inputs, n_neurons)self.biases = np.zeros((1, n_neurons))# Set regularization strengthself.weight_regularizer_l1 = weight_regularizer_l1self.weight_regularizer_l2 = weight_regularizer_l2self.bias_regularizer_l1 = bias_regularizer_l1self.bias_regularizer_l2 = bias_regularizer_l2# Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Calculate output values from inputs, weights and biasesself.output = np.dot(inputs, self.weights) + self.biases# Backward passdef backward(self, dvalues):# Gradients on parametersself.dweights = np.dot(self.inputs.T, dvalues)self.dbiases = np.sum(dvalues, axis=0, keepdims=True)# Gradients on regularization# L1 on weightsif self.weight_regularizer_l1 > 0:dL1 = np.ones_like(self.weights)dL1[self.weights < 0] = -1self.dweights += self.weight_regularizer_l1 * dL1# L2 on weightsif self.weight_regularizer_l2 > 0:self.dweights += 2 * self.weight_regularizer_l2 * self.weights# L1 on biasesif self.bias_regularizer_l1 > 0:dL1 = np.ones_like(self.biases)dL1[self.biases < 0] = -1self.dbiases += self.bias_regularizer_l1 * dL1# L2 on biasesif self.bias_regularizer_l2 > 0:self.dbiases += 2 * self.bias_regularizer_l2 * self.biases# Gradient on valuesself.dinputs = np.dot(dvalues, self.weights.T)# Dropout
class Layer_Dropout: # Initdef __init__(self, rate):# Store rate, we invert it as for example for dropout# of 0.1 we need success rate of 0.9self.rate = 1 - rate# Forward passdef forward(self, inputs):# Save input valuesself.inputs = inputs# Generate and save scaled maskself.binary_mask = np.random.binomial(1, self.rate, size=inputs.shape) / self.rate# Apply mask to output valuesself.output = inputs * self.binary_mask# Backward passdef backward(self, dvalues):# Gradient on valuesself.dinputs = dvalues * self.binary_mask# ReLU activation
class Activation_ReLU: # Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Calculate output values from inputsself.output = np.maximum(0, inputs)# Backward passdef backward(self, dvalues):# Since we need to modify original variable,# let's make a copy of values firstself.dinputs = dvalues.copy()# Zero gradient where input values were negativeself.dinputs[self.inputs <= 0] = 0# Softmax activation
class Activation_Softmax:# Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Get unnormalized probabilitiesexp_values = np.exp(inputs - np.max(inputs, axis=1, keepdims=True))# Normalize them for each sampleprobabilities = exp_values / np.sum(exp_values, axis=1, keepdims=True)self.output = probabilities# Backward passdef backward(self, dvalues):# Create uninitialized arrayself.dinputs = np.empty_like(dvalues)# Enumerate outputs and gradientsfor index, (single_output, single_dvalues) in enumerate(zip(self.output, dvalues)):# Flatten output arraysingle_output = single_output.reshape(-1, 1)# Calculate Jacobian matrix of the output andjacobian_matrix = np.diagflat(single_output) - np.dot(single_output, single_output.T)# Calculate sample-wise gradient# and add it to the array of sample gradientsself.dinputs[index] = np.dot(jacobian_matrix, single_dvalues)# Sigmoid activation
class Activation_Sigmoid:# Forward passdef forward(self, inputs):# Save input and calculate/save output# of the sigmoid functionself.inputs = inputsself.output = 1 / (1 + np.exp(-inputs))# Backward passdef backward(self, dvalues):# Derivative - calculates from output of the sigmoid functionself.dinputs = dvalues * (1 - self.output) * self.output# Linear activation
class Activation_Linear:# Forward passdef forward(self, inputs):# Just remember valuesself.inputs = inputsself.output = inputs# Backward passdef backward(self, dvalues):# derivative is 1, 1 * dvalues = dvalues - the chain ruleself.dinputs = dvalues.copy()# SGD optimizer
class Optimizer_SGD:# Initialize optimizer - set settings,# learning rate of 1. is default for this optimizerdef __init__(self, learning_rate=1., decay=0., momentum=0.):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.momentum = momentum# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If we use momentumif self.momentum:# If layer does not contain momentum arrays, create them# filled with zerosif not hasattr(layer, 'weight_momentums'):layer.weight_momentums = np.zeros_like(layer.weights)# If there is no momentum array for weights# The array doesn't exist for biases yet either.layer.bias_momentums = np.zeros_like(layer.biases)# Build weight updates with momentum - take previous# updates multiplied by retain factor and update with# current gradientsweight_updates = self.momentum * layer.weight_momentums - self.current_learning_rate * layer.dweightslayer.weight_momentums = weight_updates# Build bias updatesbias_updates = self.momentum * layer.bias_momentums - self.current_learning_rate * layer.dbiaseslayer.bias_momentums = bias_updates# Vanilla SGD updates (as before momentum update)else:weight_updates = -self.current_learning_rate * layer.dweightsbias_updates = -self.current_learning_rate * layer.dbiases# Update weights and biases using either# vanilla or momentum updateslayer.weights += weight_updateslayer.biases += bias_updates# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1 # Adagrad optimizer
class Optimizer_Adagrad:# Initialize optimizer - set settingsdef __init__(self, learning_rate=1., decay=0., epsilon=1e-7):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilon# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_cache = np.zeros_like(layer.weights)layer.bias_cache = np.zeros_like(layer.biases)# Update cache with squared current gradientslayer.weight_cache += layer.dweights**2layer.bias_cache += layer.dbiases**2# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * layer.dweights / (np.sqrt(layer.weight_cache) + self.epsilon)layer.biases += -self.current_learning_rate * layer.dbiases / (np.sqrt(layer.bias_cache) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# RMSprop optimizer
class Optimizer_RMSprop: # Initialize optimizer - set settingsdef __init__(self, learning_rate=0.001, decay=0., epsilon=1e-7, rho=0.9):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilonself.rho = rho# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_cache = np.zeros_like(layer.weights)layer.bias_cache = np.zeros_like(layer.biases)# Update cache with squared current gradientslayer.weight_cache = self.rho * layer.weight_cache + (1 - self.rho) * layer.dweights**2layer.bias_cache = self.rho * layer.bias_cache + (1 - self.rho) * layer.dbiases**2# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * layer.dweights / (np.sqrt(layer.weight_cache) + self.epsilon)layer.biases += -self.current_learning_rate * layer.dbiases / (np.sqrt(layer.bias_cache) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# Adam optimizer
class Optimizer_Adam:# Initialize optimizer - set settingsdef __init__(self, learning_rate=0.001, decay=0., epsilon=1e-7, beta_1=0.9, beta_2=0.999):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilonself.beta_1 = beta_1self.beta_2 = beta_2# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations)) # Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_momentums = np.zeros_like(layer.weights)layer.weight_cache = np.zeros_like(layer.weights)layer.bias_momentums = np.zeros_like(layer.biases)layer.bias_cache = np.zeros_like(layer.biases)# Update momentum with current gradientslayer.weight_momentums = self.beta_1 * layer.weight_momentums + (1 - self.beta_1) * layer.dweightslayer.bias_momentums = self.beta_1 * layer.bias_momentums + (1 - self.beta_1) * layer.dbiases# Get corrected momentum# self.iteration is 0 at first pass# and we need to start with 1 hereweight_momentums_corrected = layer.weight_momentums / (1 - self.beta_1 ** (self.iterations + 1))bias_momentums_corrected = layer.bias_momentums / (1 - self.beta_1 ** (self.iterations + 1))# Update cache with squared current gradientslayer.weight_cache = self.beta_2 * layer.weight_cache + (1 - self.beta_2) * layer.dweights**2layer.bias_cache = self.beta_2 * layer.bias_cache + (1 - self.beta_2) * layer.dbiases**2# Get corrected cacheweight_cache_corrected = layer.weight_cache / (1 - self.beta_2 ** (self.iterations + 1))bias_cache_corrected = layer.bias_cache / (1 - self.beta_2 ** (self.iterations + 1))# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * weight_momentums_corrected / (np.sqrt(weight_cache_corrected) + self.epsilon)layer.biases += -self.current_learning_rate * bias_momentums_corrected / (np.sqrt(bias_cache_corrected) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# Common loss class
class Loss:# Regularization loss calculationdef regularization_loss(self, layer): # 0 by defaultregularization_loss = 0# L1 regularization - weights# calculate only when factor greater than 0if layer.weight_regularizer_l1 > 0:regularization_loss += layer.weight_regularizer_l1 * np.sum(np.abs(layer.weights))# L2 regularization - weightsif layer.weight_regularizer_l2 > 0:regularization_loss += layer.weight_regularizer_l2 * np.sum(layer.weights * layer.weights)# L1 regularization - biases# calculate only when factor greater than 0if layer.bias_regularizer_l1 > 0:regularization_loss += layer.bias_regularizer_l1 * np.sum(np.abs(layer.biases))# L2 regularization - biasesif layer.bias_regularizer_l2 > 0:regularization_loss += layer.bias_regularizer_l2 * np.sum(layer.biases * layer.biases)return regularization_loss# Calculates the data and regularization losses# given model output and ground truth valuesdef calculate(self, output, y):# Calculate sample lossessample_losses = self.forward(output, y)# Calculate mean lossdata_loss = np.mean(sample_losses)# Return lossreturn data_loss# Cross-entropy loss
class Loss_CategoricalCrossentropy(Loss):# Forward passdef forward(self, y_pred, y_true):# Number of samples in a batchsamples = len(y_pred)# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valuey_pred_clipped = np.clip(y_pred, 1e-7, 1 - 1e-7)# Probabilities for target values -# only if categorical labelsif len(y_true.shape) == 1:correct_confidences = y_pred_clipped[range(samples),y_true]# Mask values - only for one-hot encoded labelselif len(y_true.shape) == 2:correct_confidences = np.sum(y_pred_clipped * y_true, axis=1)# Lossesnegative_log_likelihoods = -np.log(correct_confidences)return negative_log_likelihoods# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of labels in every sample# We'll use the first sample to count themlabels = len(dvalues[0])# If labels are sparse, turn them into one-hot vectorif len(y_true.shape) == 1:y_true = np.eye(labels)[y_true]# Calculate gradientself.dinputs = -y_true / dvalues# Normalize gradientself.dinputs = self.dinputs / samples# Binary cross-entropy loss
class Loss_BinaryCrossentropy(Loss): # Forward passdef forward(self, y_pred, y_true):# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valuey_pred_clipped = np.clip(y_pred, 1e-7, 1 - 1e-7)# Calculate sample-wise losssample_losses = -(y_true * np.log(y_pred_clipped) + (1 - y_true) * np.log(1 - y_pred_clipped))sample_losses = np.mean(sample_losses, axis=-1)# Return lossesreturn sample_losses # Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valueclipped_dvalues = np.clip(dvalues, 1e-7, 1 - 1e-7)# Calculate gradientself.dinputs = -(y_true / clipped_dvalues - (1 - y_true) / (1 - clipped_dvalues)) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Mean Squared Error loss
class Loss_MeanSquaredError(Loss): # L2 loss# Forward passdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean((y_true - y_pred)**2, axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Gradient on valuesself.dinputs = -2 * (y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Mean Absolute Error loss
class Loss_MeanAbsoluteError(Loss): # L1 lossdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean(np.abs(y_true - y_pred), axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Calculate gradientself.dinputs = np.sign(y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Softmax classifier - combined Softmax activation
# and cross-entropy loss for faster backward step
class Activation_Softmax_Loss_CategoricalCrossentropy(): # Creates activation and loss function objectsdef __init__(self):self.activation = Activation_Softmax()self.loss = Loss_CategoricalCrossentropy()# Forward passdef forward(self, inputs, y_true):# Output layer's activation functionself.activation.forward(inputs)# Set the outputself.output = self.activation.output# Calculate and return loss valuereturn self.loss.calculate(self.output, y_true)# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues) # If labels are one-hot encoded,# turn them into discrete valuesif len(y_true.shape) == 2:y_true = np.argmax(y_true, axis=1)# Copy so we can safely modifyself.dinputs = dvalues.copy()# Calculate gradientself.dinputs[range(samples), y_true] -= 1# Normalize gradientself.dinputs = self.dinputs / samples# Create dataset
X, y = sine_data()# Create Dense layer with 1 input feature and 64 output values
dense1 = Layer_Dense(1, 64)# Create ReLU activation (to be used with Dense layer):
activation1 = Activation_ReLU()# Create second Dense layer with 64 input features (as we take output
# of previous layer here) and 1 output value
dense2 = Layer_Dense(64, 1)
# Create Linear activation:
activation2 = Activation_Linear()# Create loss function
loss_function = Loss_MeanSquaredError()# Create optimizer
optimizer = Optimizer_Adam()# Accuracy precision for accuracy calculation
# There are no really accuracy factor for regression problem,
# but we can simulate/approximate it. We'll calculate it by checking
# how many values have a difference to their ground truth equivalent
# less than given precision
# We'll calculate this precision as a fraction of standard deviation
# of al the ground truth values
accuracy_precision = np.std(y) / 250# Train in loop
for epoch in range(10001):# Perform a forward pass of our training data through this layerdense1.forward(X)# Perform a forward pass through activation function# takes the output of first dense layer hereactivation1.forward(dense1.output)# Perform a forward pass through second Dense layer# takes outputs of activation function# of first layer as inputsdense2.forward(activation1.output)# Perform a forward pass through activation function# takes the output of second dense layer hereactivation2.forward(dense2.output)# Calculate the data lossdata_loss = loss_function.calculate(activation2.output, y)# Calculate regularization penaltyregularization_loss = loss_function.regularization_loss(dense1) + loss_function.regularization_loss(dense2)# Calculate overall lossloss = data_loss + regularization_loss# Calculate accuracy from output of activation2 and targets# To calculate it we're taking absolute difference between# predictions and ground truth values and compare if differences# are lower than given precision valuepredictions = activation2.outputaccuracy = np.mean(np.absolute(predictions - y) < accuracy_precision)if not epoch % 100:print(f'epoch: {epoch}, ' + f'acc: {accuracy:.3f}, ' + f'loss: {loss:.3f} (' + f'data_loss: {data_loss:.3f}, ' + f'reg_loss: {regularization_loss:.3f}), ' + f'lr: {optimizer.current_learning_rate}')# Backward passloss_function.backward(activation2.output, y)activation2.backward(loss_function.dinputs)dense2.backward(activation2.dinputs)activation1.backward(dense2.dinputs)dense1.backward(activation1.dinputs)# Update weights and biasesoptimizer.pre_update_params()optimizer.update_params(dense1)optimizer.update_params(dense2)optimizer.post_update_params()
>>>
epoch: 0, acc: 0.002, loss: 0.500 (data_loss: 0.500, reg_loss: 0.000), lr: 0.001
epoch: 100, acc: 0.003, loss: 0.346 (data_loss: 0.346, reg_loss: 0.000), lr: 0.001
...
epoch: 9900, acc: 0.003, loss: 0.145 (data_loss: 0.145, reg_loss: 0.000), lr: 0.001
epoch: 10000, acc: 0.004, loss: 0.145 (data_loss: 0.145, reg_loss: 0.000), lr: 0.001
这里的训练效果并不理想!让我们添加绘制测试数据的功能,同时对测试数据进行前向传递,在同一张图上绘制输出数据:
import matplotlib.pyplot as pltX_test, y_test = sine_data()dense1.forward(X_test)
activation1.forward(dense1.output)
dense2.forward(activation1.output)
activation2.forward(dense2.output)plt.plot(X_test, y_test)
plt.plot(X_test, activation2.output)
plt.show()
首先,我们导入了matplotlib库,然后创建了一组新的数据。接下来,有4行代码与我们之前代码中前向传播的代码相同。我们可以将其称为预测,或者在我们接下来要讨论的内容中,将其视为验证。我们将在未来的章节中解释验证和预测是什么。目前,理解我们正在做的事情就足够了:我们在与训练模型相同的特征集上进行预测,以查看模型学习到了什么,并返回了什么结果——即输出与训练时的真实目标值有多接近。然后我们绘制训练数据(显然是一个正弦波)和预测数据(我们希望其也形成一个正弦波)。让我们再次运行这段代码,并查看生成的图像:

培训过程动画:

代码可视化:https://nnfs.io/ghi
回顾一下修正线性激活函数(ReLU),它的非线性行为使我们能够映射非线性函数,但我们还需要两个或更多隐藏层。而在这里,我们只有1个隐藏层,后面是输出层。正如我们现在应该知道的那样,这显然是不够的!
如果我们再增加一层:
# Create dataset
X, y = sine_data()# Create Dense layer with 1 input feature and 64 output values
dense1 = Layer_Dense(1, 64)# Create ReLU activation (to be used with Dense layer):
activation1 = Activation_ReLU()# Create second Dense layer with 64 input features (as we take output
# of previous layer here) and 64 output values
dense2 = Layer_Dense(64, 64)# Create ReLU activation (to be used with Dense layer):
activation2 = Activation_ReLU()# Create third Dense layer with 64 input features (as we take output
# of previous layer here) and 1 output value
dense3 = Layer_Dense(64, 1)# Create Linear activation:
activation3 = Activation_Linear()# Create loss function
loss_function = Loss_MeanSquaredError()# Create optimizer
optimizer = Optimizer_Adam()# Accuracy precision for accuracy calculation
# There are no really accuracy factor for regression problem,
# but we can simulate/approximate it. We'll calculate it by checking
# how many values have a difference to their ground truth equivalent
# less than given precision
# We'll calculate this precision as a fraction of standard deviation
# of al the ground truth values
accuracy_precision = np.std(y) / 250# Train in loop
for epoch in range(10001):# Perform a forward pass of our training data through this layerdense1.forward(X)# Perform a forward pass through activation function# takes the output of first dense layer hereactivation1.forward(dense1.output)# Perform a forward pass through second Dense layer# takes outputs of activation function# of first layer as inputsdense2.forward(activation1.output)# Perform a forward pass through activation function# takes the output of second dense layer hereactivation2.forward(dense2.output)# Perform a forward pass through third Dense layer# takes outputs of activation function of second layer as inputsdense3.forward(activation2.output)# Perform a forward pass through activation function# takes the output of third dense layer hereactivation3.forward(dense3.output)# Calculate the data lossdata_loss = loss_function.calculate(activation3.output, y)# Calculate regularization penaltyregularization_loss = loss_function.regularization_loss(dense1) + loss_function.regularization_loss(dense2) + loss_function.regularization_loss(dense3)# Calculate overall lossloss = data_loss + regularization_loss# Calculate accuracy from output of activation2 and targets# To calculate it we're taking absolute difference between# predictions and ground truth values and compare if differences# are lower than given precision valuepredictions = activation3.outputaccuracy = np.mean(np.absolute(predictions - y) < accuracy_precision)if not epoch % 100:print(f'epoch: {epoch}, ' + f'acc: {accuracy:.3f}, ' + f'loss: {loss:.3f} (' + f'data_loss: {data_loss:.3f}, ' + f'reg_loss: {regularization_loss:.3f}), ' + f'lr: {optimizer.current_learning_rate}')# Backward passloss_function.backward(activation3.output, y)activation3.backward(loss_function.dinputs)dense3.backward(activation3.dinputs)activation2.backward(dense3.dinputs)dense2.backward(activation2.dinputs)activation1.backward(dense2.dinputs)dense1.backward(activation1.dinputs)# Update weights and biasesoptimizer.pre_update_params()optimizer.update_params(dense1)optimizer.update_params(dense2)optimizer.update_params(dense3)optimizer.post_update_params()import matplotlib.pyplot as pltX_test, y_test = sine_data()dense1.forward(X_test)
activation1.forward(dense1.output)
dense2.forward(activation1.output)
activation2.forward(dense2.output)
dense3.forward(activation2.output)
activation3.forward(dense3.output)plt.plot(X_test, y_test)
plt.plot(X_test, activation3.output)
plt.show()
>>>
epoch: 0, acc: 0.002, loss: 0.500 (data_loss: 0.500, reg_loss: 0.000), lr: 0.001
epoch: 100, acc: 0.003, loss: 0.187 (data_loss: 0.187, reg_loss: 0.000), lr: 0.001
...
epoch: 9900, acc: 0.617, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.001
epoch: 10000, acc: 0.620, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.001


代码可视化:https://nnfs.io/hij
我们的模型准确率并不理想,损失似乎停留在一个较高的水平。从图像中我们可以看到原因,模型在拟合数据时遇到了一些困难,看起来可能卡在了局部最小值。正如我们已经学习过的那样,为了帮助模型跳出局部最小值,我们可以尝试使用更高的学习率并添加学习率衰减。在之前的模型中,我们使用了默认的学习率0.001。现在我们将其设置为0.01,并添加学习率衰减:
optimizer = Optimizer_Adam(learning_rate=0.01, decay=1e-3)
>>>
epoch: 0, acc: 0.002, loss: 0.500 (data_loss: 0.500, reg_loss: 0.000), lr: 0.01
epoch: 100, acc: 0.027, loss: 0.061 (data_loss: 0.061, reg_loss: 0.000), lr: 0.009099181073703368
...
epoch: 9900, acc: 0.565, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.0009175153683824203
epoch: 10000, acc: 0.564, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.0009091735612328393

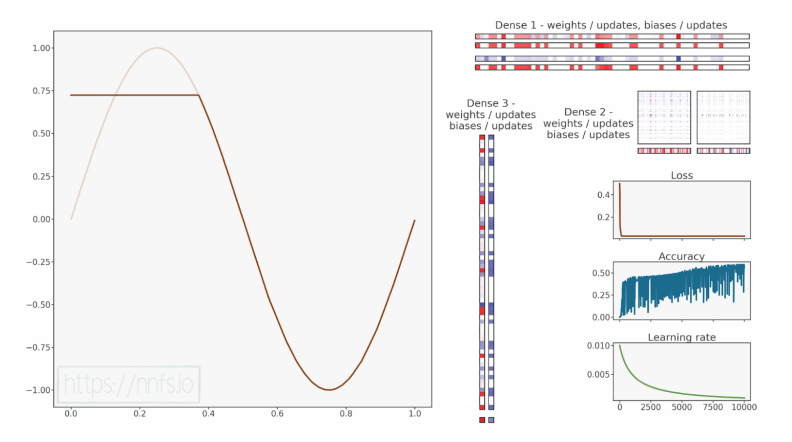
代码可视化:https://nnfs.io/ijk
这次我们的模型似乎仍然停留在更低的准确率上。那就让我们尝试使用更大的学习率吧:
optimizer = Optimizer_Adam(learning_rate=0.05, decay=1e-3)
>>>
epoch: 0, acc: 0.002, loss: 0.500 (data_loss: 0.500, reg_loss: 0.000), lr: 0.05
epoch: 100, acc: 0.087, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.04549590536851684
...
epoch: 9900, acc: 0.275, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.004587576841912101
epoch: 10000, acc: 0.229, loss: 0.031 (data_loss: 0.031, reg_loss: 0.000), lr: 0.0045458678061641965


代码可视化:https://nnfs.io/jkl
情况变得更加糟糕了。准确率显著下降,我们可以观察到正弦曲线的下部分形状也变得更差了。似乎我们无法让这个模型学习到数据,但经过多次测试和调整超参数后,我们发现学习率设置为0.005时效果更好:
optimizer = Optimizer_Adam(learning_rate=0.005, decay=1e-3)
>>>
epoch: 0, acc: 0.003, loss: 0.496 (data_loss: 0.496, reg_loss: 0.000), lr: 0.005
epoch: 100, acc: 0.017, loss: 0.048 (data_loss: 0.048, reg_loss: 0.000), lr: 0.004549590536851684
...
epoch: 9900, acc: 0.982, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.00045875768419121016
epoch: 10000, acc: 0.981, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.00045458678061641964


代码可视化:https://nnfs.io/klm
这次模型学习得相当不错,但有趣的是,较低或较高的学习率都会导致准确率较低,并且损失值卡在同一位置,而介于它们之间的学习率反而有效。
调试这种问题通常是一项相当困难的任务,且超出了本书的讨论范围。准确率和损失值表明参数的更新幅度不够大,但增加学习率反而使情况变得更糟,我们只能找到这样一个单一的点,让模型能够学习。你可能还记得,在第3章中,我们讨论了参数初始化方法以及为什么要明智地进行初始化。事实证明,在当前情况下,我们可以通过将Dense层中权重初始化的因子从0.01更改为0.1,来帮助模型学习。但你可能会问——既然学习率用于决定应用到参数的梯度幅度,为什么改变这些初始值会有帮助呢?正如你所记得的,反向传播的梯度是使用权重计算的,而学习率不会影响它。这就是为什么使用正确的权重初始化很重要,而到目前为止,我们为每个模型使用的都是相同的值。
例如,如果我们查看Keras(一个神经网络框架)的源代码,我们会发现:
def glorot_uniform(seed=None):"""Glorot uniform initializer, also called Xavier uniform initializer.It draws samples from a uniform distribution within [-limit, limit]where `limit` is `sqrt(6 / (fan_in + fan_out))`where `fan_in` is the number of input units in the weight tensorand `fan_out` is the number of output units in the weight tensor.# Argumentsseed: A Python integer. Used to seed the random generator.# ReturnsAn initializer.# ReferencesGlorot & Bengio, AISTATS 2010http://jmlr.org/proceedings/papers/v9/glorot10a/glorot10a.pdf"""return VarianceScaling(scale=1.,mode='fan_avg',distribution='uniform',seed=seed)
这段代码是Keras 2库的一部分。上述内容中真正重要的是注释部分,它描述了如何初始化权重。我们可以发现有一些重要的信息需要记住——用于乘以均匀分布抽取值的因子取决于输入数量和神经元数量,而不像我们的情况那样是一个常数。这种初始化方法称为Glorot均匀分布(Glorot Uniform)。事实上,我们(本书的作者)在自己的项目中也遇到过非常类似的问题,通过改变权重初始化的方式,使模型从完全无法学习变成能够学习的状态。
对于这个模型的目的,让我们将Dense层中权重初始化的正态分布抽取值的乘数因子改为0.1,并重新运行上述四个尝试,比较结果:
self.weights = 0.1 * np.random.randn(n_inputs, n_neurons)
并重新进行了上述所有测试:
optimizer = Optimizer_Adam()
>>>
epoch: 0, acc: 0.003, loss: 0.496 (data_loss: 0.496, reg_loss: 0.000), lr: 0.001
epoch: 100, acc: 0.005, loss: 0.114 (data_loss: 0.114, reg_loss: 0.000), lr: 0.001
...
epoch: 9900, acc: 0.869, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.001
epoch: 10000, acc: 0.883, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.001


代码可视化:https://nnfs.io/lmn
这个模型之前被卡住了,现在已经达到了很高的精度。虽然还有一些明显的瑕疵,比如这个正弦曲线的底边,但整体效果较好。
optimizer = Optimizer_Adam(learning_rate=0.01, decay=1e-3)
>>>
epoch: 0, acc: 0.003, loss: 0.496 (data_loss: 0.496, reg_loss: 0.000), lr: 0.01
epoch: 100, acc: 0.065, loss: 0.011 (data_loss: 0.011, reg_loss: 0.000), lr: 0.009099181073703368
...
epoch: 9900, acc: 0.958, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.0009175153683824203
epoch: 10000, acc: 0.949, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.0009091735612328393


代码可视化:https://nnfs.io/mno
另一个以前被卡住的模型这次训练得非常好,达到了非常高的准确率。
optimizer = Optimizer_Adam(learning_rate=0.05, decay=1e-3)
>>>
epoch: 0, acc: 0.003, loss: 0.496 (data_loss: 0.496, reg_loss: 0.000), lr: 0.05
epoch: 100, acc: 0.016, loss: 0.008 (data_loss: 0.008, reg_loss: 0.000), lr: 0.04549590536851684
...
epoch: 9000, acc: 0.802, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.005000500050005001
epoch: 9100, acc: 0.233, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004950985246063967
epoch: 9200, acc: 0.434, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004902441415825081
epoch: 9300, acc: 0.838, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.0048548402757549285
epoch: 9400, acc: 0.309, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004808154630252909
epoch: 9500, acc: 0.253, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004762358319839985
epoch: 9600, acc: 0.795, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004717426172280404
epoch: 9700, acc: 0.802, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004673333956444528
epoch: 9800, acc: 0.141, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004630058338735069
epoch: 9900, acc: 0.221, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.004587576841912101
epoch: 10000, acc: 0.631, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.0045458678061641965


代码可视化:https://nnfs.io/nop
在这种优化器设置下,“跳跃”的准确率表明学习率过大,但即便如此,该模型依然相当不错地学习到了正弦函数的形状。
optimizer = Optimizer_Adam(learning_rate=0.005, decay=1e-3)
>>>
epoch: 0, acc: 0.003, loss: 0.496 (data_loss: 0.496, reg_loss: 0.000), lr: 0.005
epoch: 100, acc: 0.017, loss: 0.048 (data_loss: 0.048, reg_loss: 0.000), lr: 0.004549590536851684
...
epoch: 9900, acc: 0.982, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.00045875768419121016
epoch: 10000, acc: 0.981, loss: 0.000 (data_loss: 0.000, reg_loss: 0.000), lr: 0.00045458678061641964


代码可视化:https://nnfs.io/opq
这些超参数再次产生了最佳结果,但差距并不大。
正如我们所看到的,这一次我们的模型在所有情况下都学会了,使用不同的学习率时都没有陷入停滞。这说明改变权重初始化对训练过程的影响是多么显著。
到此为止的全部代码:
import numpy as np
import nnfs
from nnfs.datasets import spiral_data
from nnfs.datasets import sine_datannfs.init()# Dense layer
class Layer_Dense:# Layer initializationdef __init__(self, n_inputs, n_neurons,weight_regularizer_l1=0, weight_regularizer_l2=0,bias_regularizer_l1=0, bias_regularizer_l2=0):# Initialize weights and biases# self.weights = 0.01 * np.random.randn(n_inputs, n_neurons)self.weights = 0.1 * np.random.randn(n_inputs, n_neurons)self.biases = np.zeros((1, n_neurons))# Set regularization strengthself.weight_regularizer_l1 = weight_regularizer_l1self.weight_regularizer_l2 = weight_regularizer_l2self.bias_regularizer_l1 = bias_regularizer_l1self.bias_regularizer_l2 = bias_regularizer_l2# Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Calculate output values from inputs, weights and biasesself.output = np.dot(inputs, self.weights) + self.biases# Backward passdef backward(self, dvalues):# Gradients on parametersself.dweights = np.dot(self.inputs.T, dvalues)self.dbiases = np.sum(dvalues, axis=0, keepdims=True)# Gradients on regularization# L1 on weightsif self.weight_regularizer_l1 > 0:dL1 = np.ones_like(self.weights)dL1[self.weights < 0] = -1self.dweights += self.weight_regularizer_l1 * dL1# L2 on weightsif self.weight_regularizer_l2 > 0:self.dweights += 2 * self.weight_regularizer_l2 * self.weights# L1 on biasesif self.bias_regularizer_l1 > 0:dL1 = np.ones_like(self.biases)dL1[self.biases < 0] = -1self.dbiases += self.bias_regularizer_l1 * dL1# L2 on biasesif self.bias_regularizer_l2 > 0:self.dbiases += 2 * self.bias_regularizer_l2 * self.biases# Gradient on valuesself.dinputs = np.dot(dvalues, self.weights.T)# Dropout
class Layer_Dropout: # Initdef __init__(self, rate):# Store rate, we invert it as for example for dropout# of 0.1 we need success rate of 0.9self.rate = 1 - rate# Forward passdef forward(self, inputs):# Save input valuesself.inputs = inputs# Generate and save scaled maskself.binary_mask = np.random.binomial(1, self.rate, size=inputs.shape) / self.rate# Apply mask to output valuesself.output = inputs * self.binary_mask# Backward passdef backward(self, dvalues):# Gradient on valuesself.dinputs = dvalues * self.binary_mask# ReLU activation
class Activation_ReLU: # Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Calculate output values from inputsself.output = np.maximum(0, inputs)# Backward passdef backward(self, dvalues):# Since we need to modify original variable,# let's make a copy of values firstself.dinputs = dvalues.copy()# Zero gradient where input values were negativeself.dinputs[self.inputs <= 0] = 0# Softmax activation
class Activation_Softmax:# Forward passdef forward(self, inputs):# Remember input valuesself.inputs = inputs# Get unnormalized probabilitiesexp_values = np.exp(inputs - np.max(inputs, axis=1, keepdims=True))# Normalize them for each sampleprobabilities = exp_values / np.sum(exp_values, axis=1, keepdims=True)self.output = probabilities# Backward passdef backward(self, dvalues):# Create uninitialized arrayself.dinputs = np.empty_like(dvalues)# Enumerate outputs and gradientsfor index, (single_output, single_dvalues) in enumerate(zip(self.output, dvalues)):# Flatten output arraysingle_output = single_output.reshape(-1, 1)# Calculate Jacobian matrix of the output andjacobian_matrix = np.diagflat(single_output) - np.dot(single_output, single_output.T)# Calculate sample-wise gradient# and add it to the array of sample gradientsself.dinputs[index] = np.dot(jacobian_matrix, single_dvalues)# Sigmoid activation
class Activation_Sigmoid:# Forward passdef forward(self, inputs):# Save input and calculate/save output# of the sigmoid functionself.inputs = inputsself.output = 1 / (1 + np.exp(-inputs))# Backward passdef backward(self, dvalues):# Derivative - calculates from output of the sigmoid functionself.dinputs = dvalues * (1 - self.output) * self.output# Linear activation
class Activation_Linear:# Forward passdef forward(self, inputs):# Just remember valuesself.inputs = inputsself.output = inputs# Backward passdef backward(self, dvalues):# derivative is 1, 1 * dvalues = dvalues - the chain ruleself.dinputs = dvalues.copy()# SGD optimizer
class Optimizer_SGD:# Initialize optimizer - set settings,# learning rate of 1. is default for this optimizerdef __init__(self, learning_rate=1., decay=0., momentum=0.):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.momentum = momentum# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If we use momentumif self.momentum:# If layer does not contain momentum arrays, create them# filled with zerosif not hasattr(layer, 'weight_momentums'):layer.weight_momentums = np.zeros_like(layer.weights)# If there is no momentum array for weights# The array doesn't exist for biases yet either.layer.bias_momentums = np.zeros_like(layer.biases)# Build weight updates with momentum - take previous# updates multiplied by retain factor and update with# current gradientsweight_updates = self.momentum * layer.weight_momentums - self.current_learning_rate * layer.dweightslayer.weight_momentums = weight_updates# Build bias updatesbias_updates = self.momentum * layer.bias_momentums - self.current_learning_rate * layer.dbiaseslayer.bias_momentums = bias_updates# Vanilla SGD updates (as before momentum update)else:weight_updates = -self.current_learning_rate * layer.dweightsbias_updates = -self.current_learning_rate * layer.dbiases# Update weights and biases using either# vanilla or momentum updateslayer.weights += weight_updateslayer.biases += bias_updates# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1 # Adagrad optimizer
class Optimizer_Adagrad:# Initialize optimizer - set settingsdef __init__(self, learning_rate=1., decay=0., epsilon=1e-7):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilon# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_cache = np.zeros_like(layer.weights)layer.bias_cache = np.zeros_like(layer.biases)# Update cache with squared current gradientslayer.weight_cache += layer.dweights**2layer.bias_cache += layer.dbiases**2# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * layer.dweights / (np.sqrt(layer.weight_cache) + self.epsilon)layer.biases += -self.current_learning_rate * layer.dbiases / (np.sqrt(layer.bias_cache) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# RMSprop optimizer
class Optimizer_RMSprop: # Initialize optimizer - set settingsdef __init__(self, learning_rate=0.001, decay=0., epsilon=1e-7, rho=0.9):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilonself.rho = rho# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations))# Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_cache = np.zeros_like(layer.weights)layer.bias_cache = np.zeros_like(layer.biases)# Update cache with squared current gradientslayer.weight_cache = self.rho * layer.weight_cache + (1 - self.rho) * layer.dweights**2layer.bias_cache = self.rho * layer.bias_cache + (1 - self.rho) * layer.dbiases**2# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * layer.dweights / (np.sqrt(layer.weight_cache) + self.epsilon)layer.biases += -self.current_learning_rate * layer.dbiases / (np.sqrt(layer.bias_cache) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# Adam optimizer
class Optimizer_Adam:# Initialize optimizer - set settingsdef __init__(self, learning_rate=0.001, decay=0., epsilon=1e-7, beta_1=0.9, beta_2=0.999):self.learning_rate = learning_rateself.current_learning_rate = learning_rateself.decay = decayself.iterations = 0self.epsilon = epsilonself.beta_1 = beta_1self.beta_2 = beta_2# Call once before any parameter updatesdef pre_update_params(self):if self.decay:self.current_learning_rate = self.learning_rate * (1. / (1. + self.decay * self.iterations)) # Update parametersdef update_params(self, layer):# If layer does not contain cache arrays,# create them filled with zerosif not hasattr(layer, 'weight_cache'):layer.weight_momentums = np.zeros_like(layer.weights)layer.weight_cache = np.zeros_like(layer.weights)layer.bias_momentums = np.zeros_like(layer.biases)layer.bias_cache = np.zeros_like(layer.biases)# Update momentum with current gradientslayer.weight_momentums = self.beta_1 * layer.weight_momentums + (1 - self.beta_1) * layer.dweightslayer.bias_momentums = self.beta_1 * layer.bias_momentums + (1 - self.beta_1) * layer.dbiases# Get corrected momentum# self.iteration is 0 at first pass# and we need to start with 1 hereweight_momentums_corrected = layer.weight_momentums / (1 - self.beta_1 ** (self.iterations + 1))bias_momentums_corrected = layer.bias_momentums / (1 - self.beta_1 ** (self.iterations + 1))# Update cache with squared current gradientslayer.weight_cache = self.beta_2 * layer.weight_cache + (1 - self.beta_2) * layer.dweights**2layer.bias_cache = self.beta_2 * layer.bias_cache + (1 - self.beta_2) * layer.dbiases**2# Get corrected cacheweight_cache_corrected = layer.weight_cache / (1 - self.beta_2 ** (self.iterations + 1))bias_cache_corrected = layer.bias_cache / (1 - self.beta_2 ** (self.iterations + 1))# Vanilla SGD parameter update + normalization# with square rooted cachelayer.weights += -self.current_learning_rate * weight_momentums_corrected / (np.sqrt(weight_cache_corrected) + self.epsilon)layer.biases += -self.current_learning_rate * bias_momentums_corrected / (np.sqrt(bias_cache_corrected) + self.epsilon)# Call once after any parameter updatesdef post_update_params(self):self.iterations += 1# Common loss class
class Loss:# Regularization loss calculationdef regularization_loss(self, layer): # 0 by defaultregularization_loss = 0# L1 regularization - weights# calculate only when factor greater than 0if layer.weight_regularizer_l1 > 0:regularization_loss += layer.weight_regularizer_l1 * np.sum(np.abs(layer.weights))# L2 regularization - weightsif layer.weight_regularizer_l2 > 0:regularization_loss += layer.weight_regularizer_l2 * np.sum(layer.weights * layer.weights)# L1 regularization - biases# calculate only when factor greater than 0if layer.bias_regularizer_l1 > 0:regularization_loss += layer.bias_regularizer_l1 * np.sum(np.abs(layer.biases))# L2 regularization - biasesif layer.bias_regularizer_l2 > 0:regularization_loss += layer.bias_regularizer_l2 * np.sum(layer.biases * layer.biases)return regularization_loss# Calculates the data and regularization losses# given model output and ground truth valuesdef calculate(self, output, y):# Calculate sample lossessample_losses = self.forward(output, y)# Calculate mean lossdata_loss = np.mean(sample_losses)# Return lossreturn data_loss# Cross-entropy loss
class Loss_CategoricalCrossentropy(Loss):# Forward passdef forward(self, y_pred, y_true):# Number of samples in a batchsamples = len(y_pred)# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valuey_pred_clipped = np.clip(y_pred, 1e-7, 1 - 1e-7)# Probabilities for target values -# only if categorical labelsif len(y_true.shape) == 1:correct_confidences = y_pred_clipped[range(samples),y_true]# Mask values - only for one-hot encoded labelselif len(y_true.shape) == 2:correct_confidences = np.sum(y_pred_clipped * y_true, axis=1)# Lossesnegative_log_likelihoods = -np.log(correct_confidences)return negative_log_likelihoods# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of labels in every sample# We'll use the first sample to count themlabels = len(dvalues[0])# If labels are sparse, turn them into one-hot vectorif len(y_true.shape) == 1:y_true = np.eye(labels)[y_true]# Calculate gradientself.dinputs = -y_true / dvalues# Normalize gradientself.dinputs = self.dinputs / samples# Binary cross-entropy loss
class Loss_BinaryCrossentropy(Loss): # Forward passdef forward(self, y_pred, y_true):# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valuey_pred_clipped = np.clip(y_pred, 1e-7, 1 - 1e-7)# Calculate sample-wise losssample_losses = -(y_true * np.log(y_pred_clipped) + (1 - y_true) * np.log(1 - y_pred_clipped))sample_losses = np.mean(sample_losses, axis=-1)# Return lossesreturn sample_losses # Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Clip data to prevent division by 0# Clip both sides to not drag mean towards any valueclipped_dvalues = np.clip(dvalues, 1e-7, 1 - 1e-7)# Calculate gradientself.dinputs = -(y_true / clipped_dvalues - (1 - y_true) / (1 - clipped_dvalues)) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Mean Squared Error loss
class Loss_MeanSquaredError(Loss): # L2 loss# Forward passdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean((y_true - y_pred)**2, axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Gradient on valuesself.dinputs = -2 * (y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Mean Absolute Error loss
class Loss_MeanAbsoluteError(Loss): # L1 lossdef forward(self, y_pred, y_true):# Calculate losssample_losses = np.mean(np.abs(y_true - y_pred), axis=-1)# Return lossesreturn sample_losses# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues)# Number of outputs in every sample# We'll use the first sample to count themoutputs = len(dvalues[0])# Calculate gradientself.dinputs = np.sign(y_true - dvalues) / outputs# Normalize gradientself.dinputs = self.dinputs / samples# Softmax classifier - combined Softmax activation
# and cross-entropy loss for faster backward step
class Activation_Softmax_Loss_CategoricalCrossentropy(): # Creates activation and loss function objectsdef __init__(self):self.activation = Activation_Softmax()self.loss = Loss_CategoricalCrossentropy()# Forward passdef forward(self, inputs, y_true):# Output layer's activation functionself.activation.forward(inputs)# Set the outputself.output = self.activation.output# Calculate and return loss valuereturn self.loss.calculate(self.output, y_true)# Backward passdef backward(self, dvalues, y_true):# Number of samplessamples = len(dvalues) # If labels are one-hot encoded,# turn them into discrete valuesif len(y_true.shape) == 2:y_true = np.argmax(y_true, axis=1)# Copy so we can safely modifyself.dinputs = dvalues.copy()# Calculate gradientself.dinputs[range(samples), y_true] -= 1# Normalize gradientself.dinputs = self.dinputs / samples# Create dataset
X, y = sine_data()# Create Dense layer with 1 input feature and 64 output values
dense1 = Layer_Dense(1, 64)# Create ReLU activation (to be used with Dense layer):
activation1 = Activation_ReLU()# Create second Dense layer with 64 input features (as we take output
# of previous layer here) and 64 output values
dense2 = Layer_Dense(64, 64)# Create ReLU activation (to be used with Dense layer):
activation2 = Activation_ReLU()# Create third Dense layer with 64 input features (as we take output
# of previous layer here) and 1 output value
dense3 = Layer_Dense(64, 1)# Create Linear activation:
activation3 = Activation_Linear()# Create loss function
loss_function = Loss_MeanSquaredError()# Create optimizer
# optimizer = Optimizer_Adam()
# optimizer = Optimizer_Adam(learning_rate=0.01, decay=1e-3)
# optimizer = Optimizer_Adam(learning_rate=0.05, decay=1e-3)
optimizer = Optimizer_Adam(learning_rate=0.005, decay=1e-3)# Accuracy precision for accuracy calculation
# There are no really accuracy factor for regression problem,
# but we can simulate/approximate it. We'll calculate it by checking
# how many values have a difference to their ground truth equivalent
# less than given precision
# We'll calculate this precision as a fraction of standard deviation
# of al the ground truth values
accuracy_precision = np.std(y) / 250# Train in loop
for epoch in range(10001):# Perform a forward pass of our training data through this layerdense1.forward(X)# Perform a forward pass through activation function# takes the output of first dense layer hereactivation1.forward(dense1.output)# Perform a forward pass through second Dense layer# takes outputs of activation function# of first layer as inputsdense2.forward(activation1.output)# Perform a forward pass through activation function# takes the output of second dense layer hereactivation2.forward(dense2.output)# Perform a forward pass through third Dense layer# takes outputs of activation function of second layer as inputsdense3.forward(activation2.output)# Perform a forward pass through activation function# takes the output of third dense layer hereactivation3.forward(dense3.output)# Calculate the data lossdata_loss = loss_function.calculate(activation3.output, y)# Calculate regularization penaltyregularization_loss = loss_function.regularization_loss(dense1) + loss_function.regularization_loss(dense2) + loss_function.regularization_loss(dense3)# Calculate overall lossloss = data_loss + regularization_loss# Calculate accuracy from output of activation2 and targets# To calculate it we're taking absolute difference between# predictions and ground truth values and compare if differences# are lower than given precision valuepredictions = activation3.outputaccuracy = np.mean(np.absolute(predictions - y) < accuracy_precision)if not epoch % 100:print(f'epoch: {epoch}, ' + f'acc: {accuracy:.3f}, ' + f'loss: {loss:.3f} (' + f'data_loss: {data_loss:.3f}, ' + f'reg_loss: {regularization_loss:.3f}), ' + f'lr: {optimizer.current_learning_rate}')# Backward passloss_function.backward(activation3.output, y)activation3.backward(loss_function.dinputs)dense3.backward(activation3.dinputs)activation2.backward(dense3.dinputs)dense2.backward(activation2.dinputs)activation1.backward(dense2.dinputs)dense1.backward(activation1.dinputs)# Update weights and biasesoptimizer.pre_update_params()optimizer.update_params(dense1)optimizer.update_params(dense2)optimizer.update_params(dense3)optimizer.post_update_params()import matplotlib.pyplot as pltX_test, y_test = sine_data()dense1.forward(X_test)
activation1.forward(dense1.output)
dense2.forward(activation1.output)
activation2.forward(dense2.output)
dense3.forward(activation2.output)
activation3.forward(dense3.output)plt.plot(X_test, y_test)
plt.plot(X_test, activation3.output)
plt.show()
本章的章节代码、更多资源和勘误表:https://nnfs.io/ch17
相关文章:

用 Python 从零开始创建神经网络(十七):回归(Regression)
回归(Regression) 引言1. 线性激活(Linear Activation)2. 均方误差损失(Mean Squared Error Loss)3. 均方误差损失导数(Mean Squared Error Loss Derivative)4. 平均平方误差 (MSE) …...

gentoo安装Xfce桌面
一、安装Xfce 1.选择一个配置文件 具体步骤可参见笔者的另一篇博客https://blog.csdn.net/my1114/article/details/143919066,配置文件选择24. 2.安装Xfce (1)root #emerge --ask xfce-base/xfce4-meta 第一次启动登录后时可能还需starx来启动X11 (2)安装slim&#…...

阿尔茨海默症数据集,使用yolo,voc,coco格式对2013张原始图片进行标注,可识别轻微,中等和正常的症状
阿尔茨海默症数据集,使用yolo,voc,coco格式对2013张原始图片进行标注,可识别轻微,中等,严重和正常的症状 数据集分割 训练组100% 2013图片 有效集% 0图片 测试集…...

【物联网技术与应用】实验4:继电器实验
实验4 继电器实验 【实验介绍】 继电器是一种用于响应施加的输入信号而在两个或多个点或设备之间提供连接的设备。换句话说,继电器提供了控制器和设备之间的隔离,因为设备可以在AC和DC上工作。但是,他们从微控制器接收信号,因此…...

lvs介绍与应用
LVS介绍 LVS(Linux Virtual Server)是一种基于Linux操作系统的虚拟服务器技术,主要用于实现负载均衡和高可用性。它通过将客户端请求分发到多台后端服务器上,从而提高整体服务的处理能力和可靠性。lvs是基于集群的方式实现 集群…...

Group FLUX - User Usage Survey Report
文章目录 User Feedback Summary: Software Advantages and FeaturesUser Feedback Issues and Suggested Improvements1. Security Concerns:Improvement Measures: 2. System Performance and Loading Speed:Improvement Measures: 3. Data Display Issues:Improvement Measu…...

XXE靶机攻略
XXE-Lab靶场 1.随便输入账号密码 2.使用bp抓包 3.插入xxl代码,得到结果 xxe靶机 1.安装好靶机,然后输入arp-scan -l,查找ip 2.输入ip 3.使用御剑扫描子域名 4.输入子域名 5.输入账号密码抓包 6.插入xml代码 7.使用工具解码 8.解码完毕放入文…...

第78期 | GPTSecurity周报
GPTSecurity是一个涵盖了前沿学术研究和实践经验分享的社区,集成了生成预训练Transformer(GPT)、人工智能生成内容(AIGC)以及大语言模型(LLM)等安全领域应用的知识。在这里,您可以找…...

电容Q值、损耗角、应用
电容发热的主要原因:纹波电压 当电容两端施加纹波电压时,电容承受的是变化的电压,由于电容内部存在寄生电阻(ESR)和寄生电感(ESL).因此电容会有能量损耗,从而产生热量,这…...

【WRF教程第3.6期】预处理系统 WPS 详解:以4.5版本为例
预处理系统 WPS 详解:以4.5版本为例 Geogrid/Metgrid 插值选项详解1. 插值方法的工作机制2. 插值方法的详细说明2.1 四点双线性插值(four_pt)2.2 十六点重叠抛物线插值(sixteen_pt)2.3 简单四点平均插值(av…...

linux 安装redis
下载地址 通过网盘分享的文件:redis-7.2.3.tar.gz 链接: https://pan.baidu.com/s/1KjGJB1IRIr9ehGRKBLgp4w?pwd0012 提取码: 0012 解压 tar -zxvf redis-7.2.3.tar.gz mv redis-7.2.3 /usr/local/ cd /usr/local/redis-7.2.3 安装 make install 修改配置文件 /搜索…...

Linux - rpm yum 工具及命令总结
RPM 概述 定义:RPM(RedHat Package Manager),是一个功能强大的软件包管理系统,用于在 Linux 系统中安装、升级和管理软件包采用系统:主要用于基于 RPM 的 Linux 发行版,如 Red Hat、CentOS、S…...

电子应用设计方案-58:智能沙发系统方案设计
智能沙发系统方案设计 一、引言 智能沙发作为一种融合了舒适与科技的家居产品,旨在为用户提供更加便捷、舒适和个性化的体验。本方案将详细介绍智能沙发系统的设计思路和功能实现。 二、系统概述 1. 系统目标 - 实现多种舒适的姿势调节,满足不同用户的…...

复习打卡Linux篇
目录 1. Linux常用操作命令 2. vim编辑器 3. 用户权限 4. Linux系统信息查看 1. Linux常用操作命令 基础操作: 命令说明history查看历史执行命令ls查看指定目录下内容ls -a查看所有文件 包括隐藏文件ls -l ll查看文件详细信息,包括权限类型时间大小…...

在Ubuntu 22.04 LTS中使用PyTorch深度学习框架并调用多GPU时遇到indexSelectLargeIndex相关的断言失败【笔记】
在Ubuntu 22.04 LTS系统中,已安装配置好CUDA 12.4、cuDNN 9.1.1以及PyTorch环境 export CUDA_VISIBLE_DEVICES0,1,2,3,4,5,6,7 在PyTorch深度学习框架训练调用多GPU时,提示 indexSelectLargeIndex: block: [x, 0, 0], thread: [x, 0, 0] Assertion src…...

qt 类中的run线程
在Qt中,QThread类的run()方法是线程的执行入口,它是由QThread内部自动调用的,而不是用户直接调用。 详细解释: QThread类: QThread是Qt的线程类,提供了用于多线程操作的接口。我们可以创建QThread对象并将…...

Vue3父子组件传属性和方法调用Demo
Vue3父子组件传属性和方法调用Demo 说明目录父组件给子组件传值和方法 父组件给子组件传值-使用defineProps接受父组件属性值父组件给子组件传值-使用defineModel接受父组件v-model值 当子组件只需要接收父组件一个v-model值时,写法1如下:子组件接收单个v-model写法2如下:当子…...
aac怎么转为mp3?操作起来很简单的几种aac转mp3的方法
aac怎么转为mp3?aac格式的优势主要体现在音质和压缩效率,尤其是在较低比特率下,能够实现更清晰的音质,这也是为何许多现代设备和应用偏爱aac格式的原因之一。特别是在手机、平板以及智能音响等设备中,aac文件几乎可以无…...

结合mybatis-plus实现Function获取java实体类的属性名
1、工具类 package com.yh.tunnel.util;import com.baomidou.mybatisplus.core.toolkit.support.SFunction; import com.google.common.base.CaseFormat; import com.yh.tunnel.domain.Plan;import java.lang.invoke.SerializedLambda; import java.lang.reflect.Field; import…...

vue 响应式数据原理
发现宝藏 前些天发现了一个巨牛的人工智能学习网站,通俗易懂,风趣幽默,忍不住分享一下给大家。【宝藏入口】。 Vue 的响应式数据原理是其核心功能之一,它使得 Vue 应用能够自动响应数据的变化,并在数据变化时自动更新…...

Objective-C常用命名规范总结
【OC】常用命名规范总结 文章目录 【OC】常用命名规范总结1.类名(Class Name)2.协议名(Protocol Name)3.方法名(Method Name)4.属性名(Property Name)5.局部变量/实例变量(Local / Instance Variables&…...
详解:相对定位 绝对定位 固定定位)
css的定位(position)详解:相对定位 绝对定位 固定定位
在 CSS 中,元素的定位通过 position 属性控制,共有 5 种定位模式:static(静态定位)、relative(相对定位)、absolute(绝对定位)、fixed(固定定位)和…...
)
【RockeMQ】第2节|RocketMQ快速实战以及核⼼概念详解(二)
升级Dledger高可用集群 一、主从架构的不足与Dledger的定位 主从架构缺陷 数据备份依赖Slave节点,但无自动故障转移能力,Master宕机后需人工切换,期间消息可能无法读取。Slave仅存储数据,无法主动升级为Master响应请求ÿ…...
dify打造数据可视化图表
一、概述 在日常工作和学习中,我们经常需要和数据打交道。无论是分析报告、项目展示,还是简单的数据洞察,一个清晰直观的图表,往往能胜过千言万语。 一款能让数据可视化变得超级简单的 MCP Server,由蚂蚁集团 AntV 团队…...
力扣热题100 k个一组反转链表题解
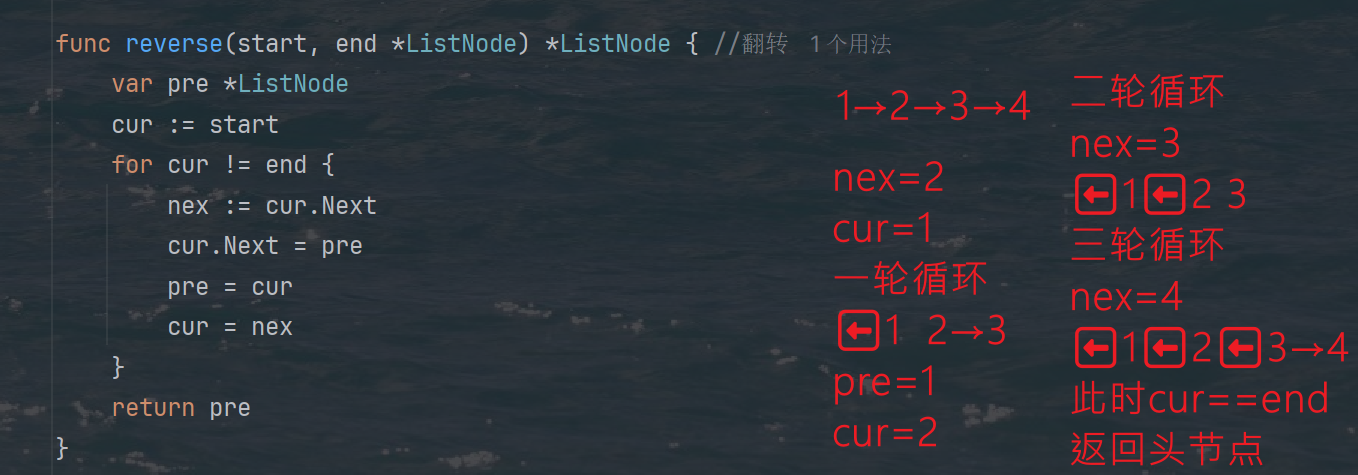
题目: 代码: func reverseKGroup(head *ListNode, k int) *ListNode {cur : headfor i : 0; i < k; i {if cur nil {return head}cur cur.Next}newHead : reverse(head, cur)head.Next reverseKGroup(cur, k)return newHead }func reverse(start, end *ListNode) *ListN…...

【Android】Android 开发 ADB 常用指令
查看当前连接的设备 adb devices 连接设备 adb connect 设备IP 断开已连接的设备 adb disconnect 设备IP 安装应用 adb install 安装包的路径 卸载应用 adb uninstall 应用包名 查看已安装的应用包名 adb shell pm list packages 查看已安装的第三方应用包名 adb shell pm list…...

MySQL 索引底层结构揭秘:B-Tree 与 B+Tree 的区别与应用
文章目录 一、背景知识:什么是 B-Tree 和 BTree? B-Tree(平衡多路查找树) BTree(B-Tree 的变种) 二、结构对比:一张图看懂 三、为什么 MySQL InnoDB 选择 BTree? 1. 范围查询更快 2…...
【LeetCode】算法详解#6 ---除自身以外数组的乘积
1.题目介绍 给定一个整数数组 nums,返回 数组 answer ,其中 answer[i] 等于 nums 中除 nums[i] 之外其余各元素的乘积 。 题目数据 保证 数组 nums之中任意元素的全部前缀元素和后缀的乘积都在 32 位 整数范围内。 请 不要使用除法,且在 O…...
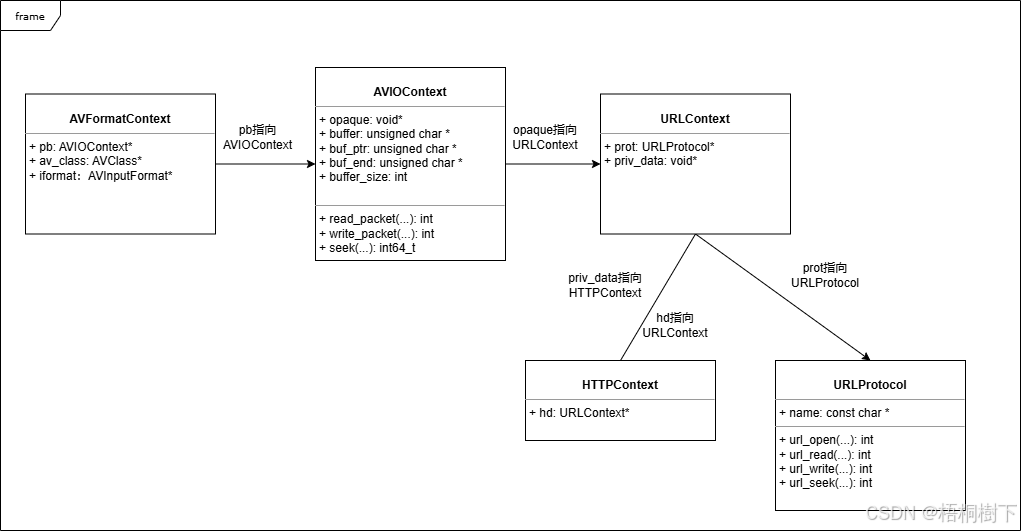
FFmpeg avformat_open_input函数分析
函数内部的总体流程如下: avformat_open_input 精简后的代码如下: int avformat_open_input(AVFormatContext **ps, const char *filename,ff_const59 AVInputFormat *fmt, AVDictionary **options) {AVFormatContext *s *ps;int i, ret 0;AVDictio…...
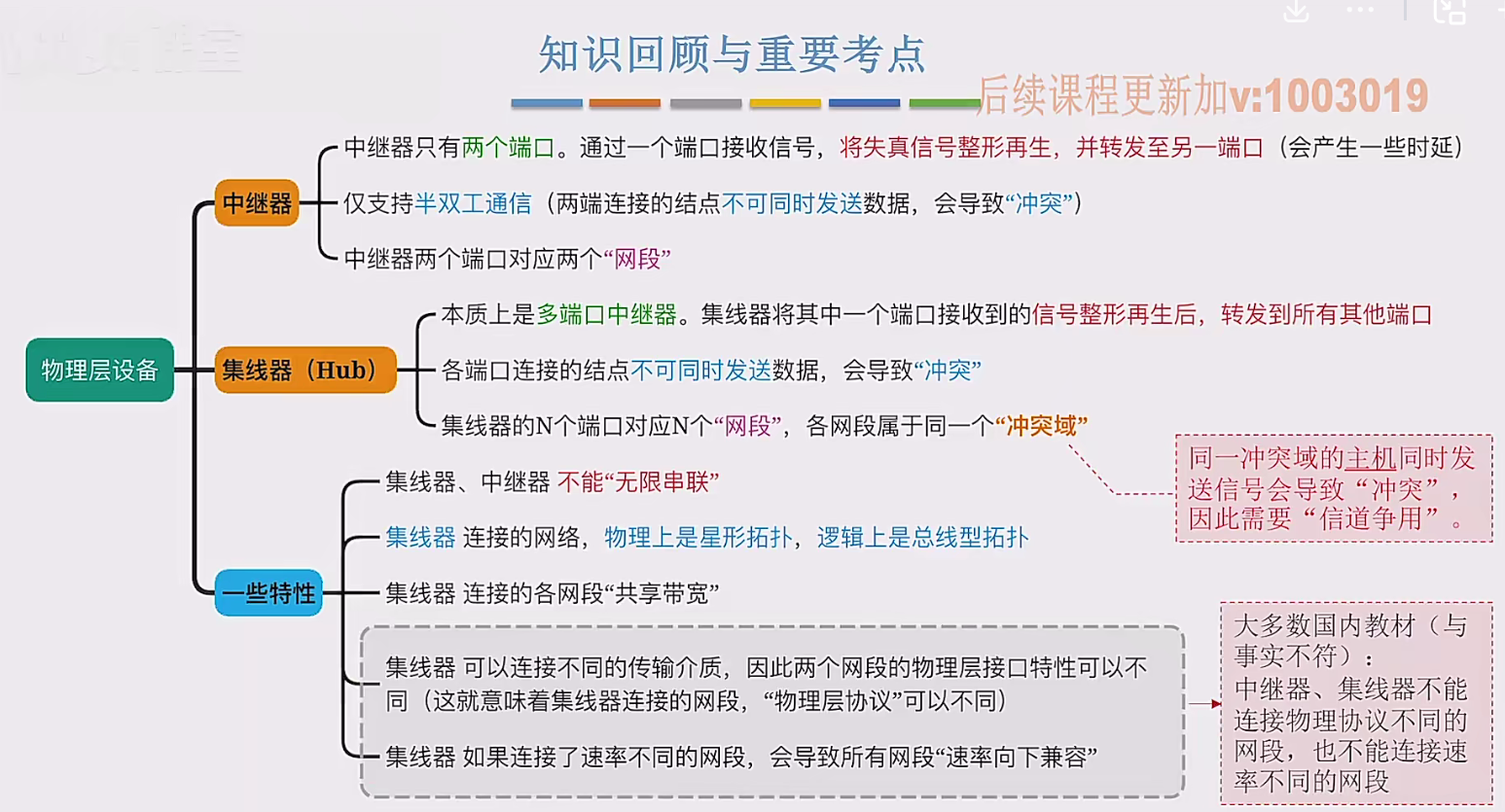
2.3 物理层设备
在这个视频中,我们要学习工作在物理层的两种网络设备,分别是中继器和集线器。首先来看中继器。在计算机网络中两个节点之间,需要通过物理传输媒体或者说物理传输介质进行连接。像同轴电缆、双绞线就是典型的传输介质,假设A节点要给…...
